Signal transduction: Difference between revisions
No edit summary |
No edit summary |
||
| Line 88: | Line 88: | ||
This large and diverse class of steroids are biosynthesized from isoprenoids and structurally resemble cholesterol. Mammalian steroid hormones can be grouped into five groups by the receptors to which they bind: glucocorticoids, mineralocorticoids, androgens, estrogens, and progestogens. Vitamin D derivatives are a sixth closely related hormone system with homologous receptors. They have some of the characteristics of true steroids as receptor ligands. For example, <scene name='89/895670/Cv/1'>estradiol</scene> is an important estrogen steroid hormone in both women and men. It is a typical steroid with core four-ring system (ABCD), composed of 17 carbon atoms. | This large and diverse class of steroids are biosynthesized from isoprenoids and structurally resemble cholesterol. Mammalian steroid hormones can be grouped into five groups by the receptors to which they bind: glucocorticoids, mineralocorticoids, androgens, estrogens, and progestogens. Vitamin D derivatives are a sixth closely related hormone system with homologous receptors. They have some of the characteristics of true steroids as receptor ligands. For example, <scene name='89/895670/Cv/1'>estradiol</scene> is an important estrogen steroid hormone in both women and men. It is a typical steroid with core four-ring system (ABCD), composed of 17 carbon atoms. | ||
''[[Corticosteroids]]'' | |||
Corticosteroids are a class of steroid hormones that are produced in the adrenal cortex of vertebrates, as well as the synthetic analogues of these hormones. Two main classes of corticosteroids, [[glucocorticoids]] and [[mineralocorticoids]], are involved in a wide range of physiological processes. | Corticosteroids are a class of steroid hormones that are produced in the adrenal cortex of vertebrates, as well as the synthetic analogues of these hormones. Two main classes of corticosteroids, [[glucocorticoids]] and [[mineralocorticoids]], are involved in a wide range of physiological processes. | ||
| Line 98: | Line 98: | ||
<scene name='89/895670/Cv/2'>Cortisol</scene> (hydrocortisone) is a corticosteroid with both glucocorticoid and mineralocorticoid activity and effects. | <scene name='89/895670/Cv/2'>Cortisol</scene> (hydrocortisone) is a corticosteroid with both glucocorticoid and mineralocorticoid activity and effects. | ||
''[[Glucocorticoids]]'' | |||
Glucocorticoids are corticosteroids that bind to the glucocorticoid receptor. <scene name='89/895670/Cv/3'>Dexamethasone</scene> is a glucocorticoid medication. It is the most potent glucocorticoid and it has not mineralocorticoid potency. | Glucocorticoids are corticosteroids that bind to the glucocorticoid receptor. <scene name='89/895670/Cv/3'>Dexamethasone</scene> is a glucocorticoid medication. It is the most potent glucocorticoid and it has not mineralocorticoid potency. | ||
*[[Glucocorticoid receptor]]. <scene name='89/895670/Cv/4'>Human glucocorticoid receptor ligand-binding domain bound to dexamethasone</scene> ([[1m2z]]). | *[[Glucocorticoid receptor]]. <scene name='89/895670/Cv/4'>Human glucocorticoid receptor ligand-binding domain bound to dexamethasone</scene> ([[1m2z]]). | ||
| Line 117: | Line 117: | ||
*Microsomal [[Prostaglandin E synthase]] (PGES) converts cyclooxygenase (COX)-derived prostaglandin to PGE2. It is membrane-associated and belongs to the microsomal glutathione S-transferase family. PGES is preferentially linked with the inducible COX-2<ref>PMID:12432931</ref> . PGES is induced by proinflammatory stimuli and down-regulated by anti-inflammatory '''glucocorticoids'''<ref>PMID:16336776</ref>. Microsomal ''Prostaglandin E synthase'' <scene name='77/778890/Cv/2'>is membrane-associated</scene> (coordinates are from [http://opm.phar.umich.edu/protein.php?extrapdb=4yl0 OPM database]. The <scene name='77/778890/Cv/6'>anti-inflammatory inhibitor binds to PGES in a pocket above the glutathione and interacts with various side-chains of a helix</scene><ref>PMID:25961169</ref>. Water molecules are shown as red spheres. | *Microsomal [[Prostaglandin E synthase]] (PGES) converts cyclooxygenase (COX)-derived prostaglandin to PGE2. It is membrane-associated and belongs to the microsomal glutathione S-transferase family. PGES is preferentially linked with the inducible COX-2<ref>PMID:12432931</ref> . PGES is induced by proinflammatory stimuli and down-regulated by anti-inflammatory '''glucocorticoids'''<ref>PMID:16336776</ref>. Microsomal ''Prostaglandin E synthase'' <scene name='77/778890/Cv/2'>is membrane-associated</scene> (coordinates are from [http://opm.phar.umich.edu/protein.php?extrapdb=4yl0 OPM database]. The <scene name='77/778890/Cv/6'>anti-inflammatory inhibitor binds to PGES in a pocket above the glutathione and interacts with various side-chains of a helix</scene><ref>PMID:25961169</ref>. Water molecules are shown as red spheres. | ||
''[[Mineralocorticoids]]'' | |||
Mineralocorticoids are a class of corticosteroids. Mineralocorticoids are produced in the adrenal cortex and influence salt and water balances (electrolyte balance and fluid balance). The primary mineralocorticoid is <scene name='89/896192/Cv/1'>aldosterone</scene>. | Mineralocorticoids are a class of corticosteroids. Mineralocorticoids are produced in the adrenal cortex and influence salt and water balances (electrolyte balance and fluid balance). The primary mineralocorticoid is <scene name='89/896192/Cv/1'>aldosterone</scene>. | ||
*[[Mineralocorticoid receptor]] (MR) in epithelial cells is activated by the mineralocorticoid hormone aldosterone promoting renal sodium retention and potassium excretion. It is [[Nuclear receptors|nuclear receptor]]. In non epithelial cells MR is activated by cortisol<ref>PMID:15199296</ref>. MR is exposed to many steroids including cortisol, cortisone and progesterone, however, aldosterone and deoxycorticosterone are its physiological ligands. MR mutations are the principal cause of renal pseudohypoaldosteronism<ref>PMID:16972228</ref>. MR mutation S810L causes early-onset hypertension<ref>PMID:10884226</ref>. Inhibition of cardia MR prevents doxorubicin-induced cardiotoxicity<ref>PMID:28430882</ref>. MR is an important proadipogenic transcription factor that may mediate aldosterone and glucocorticoid effects on adipose tissue development and hence on obesity and development of metabolic syndrome<ref>PMID:17384139</ref>. The MR ligand aldosterone binds in a <scene name='78/781019/Cv/6'>fully enclosed pocket, contacting residues with six α-helices and a β-turn</scene> ({{Template:ColorKey_Helix}},{{Template:ColorKey_Strand}},{{Template:ColorKey_Loop}},{{Template:ColorKey_Turn}}). <scene name='78/781019/Cv/7'>It forms hydrogen bonds with 4 MR residues</scene><ref>PMID:15967794</ref>. <scene name='78/781019/Cv/8'>Whole binding site</scene>. Water molecules are shown as red spheres. | *[[Mineralocorticoid receptor]] (MR) in epithelial cells is activated by the mineralocorticoid hormone aldosterone promoting renal sodium retention and potassium excretion. It is [[Nuclear receptors|nuclear receptor]]. In non epithelial cells MR is activated by cortisol<ref>PMID:15199296</ref>. MR is exposed to many steroids including cortisol, cortisone and progesterone, however, aldosterone and deoxycorticosterone are its physiological ligands. MR mutations are the principal cause of renal pseudohypoaldosteronism<ref>PMID:16972228</ref>. MR mutation S810L causes early-onset hypertension<ref>PMID:10884226</ref>. Inhibition of cardia MR prevents doxorubicin-induced cardiotoxicity<ref>PMID:28430882</ref>. MR is an important proadipogenic transcription factor that may mediate aldosterone and glucocorticoid effects on adipose tissue development and hence on obesity and development of metabolic syndrome<ref>PMID:17384139</ref>. The MR ligand aldosterone binds in a <scene name='78/781019/Cv/6'>fully enclosed pocket, contacting residues with six α-helices and a β-turn</scene> ({{Template:ColorKey_Helix}},{{Template:ColorKey_Strand}},{{Template:ColorKey_Loop}},{{Template:ColorKey_Turn}}). <scene name='78/781019/Cv/7'>It forms hydrogen bonds with 4 MR residues</scene><ref>PMID:15967794</ref>. <scene name='78/781019/Cv/8'>Whole binding site</scene>. Water molecules are shown as red spheres. | ||
| Line 124: | Line 124: | ||
*[[Hydroxysteroid dehydrogenase]] | *[[Hydroxysteroid dehydrogenase]] | ||
*[[ | =Sex steroids= | ||
*[[ | ==[[Androgens]]== | ||
An androgen is any natural or synthetic steroid hormone that regulates the development and maintenance of male characteristics in vertebrates by binding to androgen receptors. The major androgen in males is <scene name='89/895670/Cv/5'>testosterone</scene>. It is the primary sex hormone and anabolic steroid in males. It is a steroid from the androstane class. It exerts its action through binding to and activation of the [[androgen receptor]]. | |||
*[[Androgen receptor]]. Ligand binding domain (LBD) containing an <scene name='54/543362/Cv/3'>active site</scene> which binds intramolecularly the N-terminal FXXFL motif or coactivators with the same motif.<ref>PMID:18805694</ref> Water molecules are shown as red spheres. <scene name='89/895670/Cv/6'>Human androgen receptor bound to testosterone</scene> ([[2ylo]]). | |||
*[[Heat shock factor]] (HSF) are transcriptional activators of heat shock genes. HSF bind to heat shock sequence elements throughout the genome with a consensus array of three oppositely oriented sequence AGGAN and activate transcription. Each HSF monomer contains one C-terminal and 3 N-terminal leucine zippers. Two sequences flanking the N-terminal leucine zippers contain the consensus nuclear localization signal (NLS). The DNA-binding domain (DBD residues 193-281) of HSF lies in the N-terminal of the first NLS region<ref>PMID:8441385</ref>. Depletion of HSF-1 is associated with accumulation of pathogenic [[androgen receptor]] in neurodegenerative diseases<ref>PMID:23360996</ref>. | |||
*[[Cellular retinoic acid-binding protein]] (CRABP); Epididymal RABP (ERABP) is an '''androgen'''-dependent RABP present in the lumen of the epididymis believed to be involved in '''sperm''' maturation. ERABP binds specifically all-trans- and 9-cis-RA. | |||
*[[Aromatase]]. The primary function of aromatase is to produce estrogens by aromatizing '''androgens'''. Aromatase is the only known enzyme in vertebrates capable of catalyzing the aromatization of a six-membered ring<ref name="structure"> Ghosh, D., Griswold, J., Erman, M., Pangborn, W. " X-ray Structure of Human Aromatase Reveals An Androgen-Specific Active Site" ''Journal of Steroid Biochemistry and Molecular Biology''. [Online] '''2010''',Vol. 118, Issue 4-5, p197-202[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2826573/]</ref>. | |||
*[[Student Project 1 for UMass Chemistry 423 Spring 2015]]. Protein kinase C related kinase 1 (PRK1) is a component of Rho-GTPase, histone demethylase, '''androgen''' receptor, and histone demethylase signaling pathways and is involved in ovary and prostate cancer. A lot of PRK1 is expressed in cases of ovarian serous carcinoma. | |||
*[[Finasteride]] | |||
*[[Zolinza (Vorinostat)]] | |||
*[[Hydroxysteroid dehydrogenase]], 17-β HSD is involved in the conversion of androstenedione to testosterone. | |||
*[[Aromatase]] converts androstenedione to estrogen and testosterone to estradiol. | |||
*[[Lipids: structure and classification]] | |||
*Cytochrome P450 3A4 ([[CYP3A4]]) | |||
==[[Estrogens]]== | |||
There are three major endogenous estrogens that have estrogenic hormonal activity: estrone (E1), estradiol (E2), and estriol (E3). Estradiol, an estrane, is the most potent and prevalent. Another estrogen called estetrol (E4) is produced only during pregnancy. | |||
*[[Estrogen receptor]] | |||
<scene name='Estrogen_receptor/Cv/1'>Click here to see the difference between conformations</scene> of estrogen receptor α complexed with raloxifene and a corepressor peptide (morph was taken from [http://molmovdb.org/cgi-bin/movie.cgi Gallery of Morphs] of the [http://molmovdb.org Yale Morph Server]). | |||
Structure of estradiol metal chelate and estrogen receptor complex: The basis for designing a new class of SERMs<ref>PMID: 21473635</ref>. | |||
Selective estrogen receptor modulators, such as estradiol 17-derived metal complexes, have been synthesized as targeted probes for the diagnosis and treatment of breast cancer. The detailed 3D structure of <scene name='Journal:JMEDCHEM:1/Cv/11'>estrogen receptor α ligand-binding domain (ER-LBD)</scene> bound with a novel <scene name='Journal:JMEDCHEM:1/Cv/5'>estradiol-derived metal complex, estradiol-pyridinium tetra acetate europium (III) (EPTA-Eu)</scene> at 2.6Å resolution was reported ([[2yat]]). The residues <scene name='Journal:JMEDCHEM:1/Cv/10'>Glu353, Arg394 and His524 and the conserved water molecule (W1006) form hydrogen bonds</scene> with EPTA-Eu. The hydrogen bonds are shown as white dashed lines. <scene name='Journal:JMEDCHEM:1/Cv/7'>Superposition</scene> of this structure with the structure of native ligand 17β-estradiol (E2) in the complex of E2/ERα-LBD complex ([[1ere]]) reveals that the <scene name='Journal:JMEDCHEM:1/Cv/12'>E2 core of EPTA-Eu overlaps closely with that of E2 itself</scene>. The <scene name='Journal:JMEDCHEM:1/Cv/9'>hydrogen bonds network</scene> made by additional estrogen receptor residues (''e.g.'' Glu419 of H7 and Glu339 of H3, this depends on subunit), may work together with the E2 17β hydroxyl-His524 hydrogen bond and tighten the neck of the LBP upon binding of the endogenous ligand E2. 4-Hydroxytamoxifen (OHT) is an other selective estrogen receptor modulator. <scene name='Journal:JMEDCHEM:1/Al/5'>Superposition</scene> of EPTA-Eu/ERα-LBD complex on OHT/ERα-LBD complex ([[3ert]]) shows that there is similar network of hydrogen bonds in both complexes, except for His524 which does not form hydrogen bond with OHT in the OHT/ERα-LBD complex. <scene name='Journal:JMEDCHEM:1/Al1/3'>Superposition of structures of all these three complexes:</scene> E2/ERα-LBD ([[1ere]]), OHT/ERα-LBD ([[3ert]]) and EPTA-Eu/ERα-LBD shows that they overlap well in the majority portions of the domain, but differ significantly in the region of the 'omega loop'. They display different synergistic reciprocating movements, depending on the specific nature of the ligand bound. The structure of estrogen receptor complexed with EPTA-Eu provides important information pertinent to the design of novel functional ER targeted probes for clinical applications. | |||
*[[Ivan Koutsopatriy estrogen receptor]] | |||
ER is a modular protein composed of a ligand binding domain, a DNA binding domain and a transactivation domain. ER is a DNA-binding transcription factor. | |||
<scene name='71/714947/Er_bound_to_dna/4'>ER bound to DNA</scene>. The DNA binding domain can be clearly observed in this scene; the highlighted yellow helix in close proximity to the DNA is part of the DNA binding domain. The blue beta sheet close to the yellow DNA binding alpha helix is also part of the DNA binding domain. The transactivation domain forms an alpha helix which is colored in purple. The transactivation domain activates RNA polymerase when the receptor binds to DNA. The ligand binding domain may be observed here with the following scene. | |||
<scene name='71/714947/Agonist_ferutinine_bound_er/5'>Agonist ferutinine bound ER</scene>. The ligand ferutinine (highlighted in pink) is bound by the ligand binding domain, composed of the blue colored alpha helices immediately surrounding the purple ligand. Another view of the ligand binding domain is shown here, with estradiol bound. <scene name='71/714947/Er_ligand_binding_domain_estra/1'>ER ligand binding domain bound to estradiol</scene>. | |||
ER is functional as a ligand-dependent transcription factor. ER responds to both agonist and antagonist ligands and can associate with the nuclear matrix. Differences in the structure of the receptor are observed depending on what ligand ER has bound (if any). Through comparisons of ER bound to agonist and antagonist ligands, some structural components may be highlighted. <scene name='71/714947/Agonist_estradiol_bound_er/2'>Agonist estradiol bound er</scene> The specific conformation of this tight loop of alpha helices and beta sheets around the ligand shows a complex capable of activating ER's transcription loci. This complex allows for the activation signal that will stimulate normal growth. | |||
Normal growth is stimulated when an agonist bound ER binds DNA. This occurs with the assistance of chaperon proteins. These chaperons are capable of recognizing estrogen receptor ligand complexes. When ER has bound a ligand chaperons facilitate the trans-location of the complex to the nucleus. Eventually the chaperon ligand ER complex will reach specific euchromatin, at which point the chaperons facilitate the ligand ER complex to changes conformation. This conformation will facilitate the estrogen receptor to bind the DNA major groove at specific palindromic sequences. Estradiol is a normal ligand for ER and allows for binding in the major groove of DNA. | |||
If the ligand is an antagonist the transcription factor function of estrogen receptor becomes hindered. <scene name='71/714947/Partial_agonist_genistein_er/3'>Partial Agonist genistein bound ER</scene> The conformation of ER bound to the partial agonist genistein has a loop which is not as tight around the ligand as those found on ER with a complete agonist ligand. The ligands themselves take up different amounts of space and have varying interactions within ER. This slight difference effects the ability of the chaperon to be able to bind the receptor ligand complex to the major groove of DNA. There is a noticeable difference in the size of the pure agonist vs partial agonist scenes. Specifically, look at the width of the agonist compared to the partial agonist. | |||
Similar differences may be observed between ER which has bound the partial agonist and complete antagonist ligands. <scene name='71/714947/Antagonist_tamoxifen_bound_er/5'>Antagonist tamoxifen bound ER</scene> The most drastic difference is noticeable between agonist and antagonist ligands. Compare the agonist scene to the <scene name='71/714947/Agonist_estradiol_bound_er/2'>Agonist estradiol bound er</scene>. Special attention should be given to the bottom right alpha helices and beta sheets that are pushed out more in the antagonist compared to the agonist bound ER. | |||
*[[Student Project 10 for UMass Chemistry 423 Spring 2015|Estrogen receptor beta/genistein complex]] | |||
*[[Sandbox Reserved 433|Estrogen receptor beta/p-hydroxybenzene sulfonamide complexes]] | |||
<scene name='48/483891/Initial_view/1'>Estrogen receptor β</scene> (ER-β) is 1 of the 2 isoforms of the estrogen receptor, a ligand-activated transcription factor which regulates the biological effects of the steroid hormone 17 β-estradiol, or estrogen, in both males and females. The complex is a hetero-tetrameric assembly consisting of 4 molecules and a ligand: 2 copies of <scene name='48/483891/Erbeta/1'>estrogen receptor β</scene>, 2 copies of <scene name='48/483891/Steroid_receptor/3'>steroid receptor coactivator-1</scene>, and the ligand, <scene name='48/483891/Ligand/3'>Genistein</scene>. Once the ligand is bound, the complex recruits the steroid receptor coactivators, which recruit other proteins to form the transcriptional complex for initiation of transcription. This activates expression of reporter genes containing estrogen response elements. Genistein is a phytoestrogen with structural similarity to estrogen which competes for estrogen receptors. | |||
Although estrogen receptor β is widely expressed, it is not the primary estrogen receptor in most tissues. As a result, it has become a target for drug delivery, especially since it is 40x more selective for genistein than the α isoform. This enhanced selectivity may be caused by differences in residues <scene name='48/483891/Met336_ile373/2'>336 and 373</scene> between the 2 isoforms, allowing ER-β to accommodate more polar substituents in its binding pocket. ER-β differs greatly from ER-α at the N-terminal domains, which can be seen located at opposite ends from the C termini in this <scene name='48/483891/Rainbow/1'>rainbow representation</scene>. The protein is composed of 3 sections: a modulating N-terminal domain, a DNA-binding domain and a C-terminal ligand-binding domain. | |||
{{Template:ColorKey_N2CRainbow}} | |||
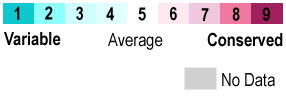
Each ERβ contains several domains with specific functions: an N-terminal domain (NTD), a {{Template:ColorKey Composition DNA}}-binding domain (DBD), a flexible hinge region and a C-terminal {{Template:ColorKey Composition Ligand}}-binding domain (LBD). The complex overall is about <scene name='48/483891/Erhelices/1'>66% helical (10 helices; 160 residues) and 3% β-sheet (2 strands; 9 residues)</scene>. The <scene name='48/483891/Ntd-erbeta/2'>NTD</scene> is the 1st activation function (AF-1) domain that consists mostly of random coils and a small portion of helices (red) and sheets (green); it is a <scene name='48/483891/Sequence_conservation/1'>variable region</scene>. This lack of structure allows the region to recruit and bond many different interaction partners. This region also has the capacity to transactivate transcription without binding estrogen. | |||
{{Template:ColorKey_ConSurf_NoYellow}} | |||
The <scene name='48/483891/Dbdlbd/1'>DBD</scene> binds estrogen response elements (ERE) of target genes and recruits coactivator proteins responsible for the transcription of these genes. The ERE consist of a palindromic inverted repeat 5'GGTCAnnnTGACC-3' of target genes. The DBD is a highly <scene name='48/483891/Sequence_conservation/1'>conserved region</scene>. It is composed of 2 C4-type Zn fingers each containing <scene name='48/483891/Dbd-erbeta/4'>4 Cys</scene> residues coordinating to a Zn atom. | |||
The hinge region connects the DBD and LBD. | |||
<scene name='48/483891/Dbdlbd/1'>LBD</scene> binds estrogen, coregulatory proteins, corepressors and coactivators. Genistein is not generated by the endocrine system that binds ERβ like estrogen; both ligands are completely buried within the <scene name='48/483891/Hydrophobic_pocket/3'>hydrophobic core</scene> | |||
({{Template:ColorKey_Hydrophobic}}, {{Template:ColorKey_Polar}}) of the ERβ complex. | |||
Binding at the LBD activates transcription mediated by the DBD. This domain is entirely helical; the LBD interacts with genistein through helices. The conformationally dynamic portion of this region gives rise to ERβ’s ligand-dependent transcriptional activation (AF-2) function. A key element of AF-2 is helix 12 (H12), which acts as a conformational switch; different receptor ligands influence the orientation of H12. Agonist ligands like genistein position H12 across the ligand-binding pocket of the LBD, which provides a coactivator docking surface. Geinstein binding allows the helices of AF-2 to form a shallow hydrophobic binding site for leucine-rich motifs of coactivators to bind. This conformation provides optimal interaction with coactivators and transcription is activated. | |||
Genistein's bicyclic form allows it to hydrogen bond on opposite sides with the hydroxyls of the histidine groups on the receptor. <scene name='48/483891/Estrogen_kyle/12'>His475's</scene> binding to the receptor causes a conformational change and activates the receptor resulting in up-regulation for coactivators. Down-regulation will occur in the presence of corepressor as they bind to repressors and indirectly regulate gene expression. In order for the estrogen receptor β genistein to bind to a receptor and activate it there must be stabilization by a coactivator. The coactivator increases the gene expression and with this increase allows it to bind to an activator group consisting of a DNA binding domain. The estrogen receptor is found to be comprised of a dimer attached to a ligand and coactivator peptide which helps to stabilize the structure of each monomer. The conformational state of helix-12 can be modified by the binding of the coactivator. | |||
This <scene name='48/483891/Estrogen_kyle/8'>scene</scene> depicts the hydrophobic and hydrophilic residues of the estrogen receptor. The hydrophobic regions are primarily on the inside of the protein surrounding genistein (red). Having the hydrophobic residues surrounding the binding pocket will stabilize the structure. The structure of this pocket is tertiary and do to the hydrophobic interactions inside the pocket and hydrophilic interactions on the outside help to stabilize this tertiary structure. The <scene name='48/483891/Estrogen_kyle/16'>binding pocket</scene> is hydrophobic which means that an increase in lipophilicity would increase the affinity for ligands which in this case is genistein. The genistein structure has 3 hydroxyl groups, an ether and an ester. These 3 functional groups are polar and have many possibilities for hydrogen bonding. The His475 and Met336 residues in the binding pocket are capable of forming hydrogen bonds with genistein do to the many hydrogen bond forming functional groups. These residues are different from the residues found in ERα and so the selectivity of genistein is much greater for ERβ. | |||
Upon visualizing the estrogen receptor in an <scene name='48/483891/Arrow_view/1'>arrow representation</scene>, the structure can be classified as parallel or anti-parallel. Here is the zoomed <scene name='48/483891/Hydrophobic_pocket/3'>primarily hydrophobic pocket</scene>. | |||
*[[Estrogen-related receptor]] | |||
*[[Tamoxifen|Tamoxifen and the Estrogen Receptor/Tamoxifen and the Estrogen-related receptor]] | |||
<scene name='50/501401/Cv/4'>Binding of nuclear receptor corepressor 2 peptide and 4-hydroxytamoxifen</scene> to human estrogen-related receptor γ. The chemotherapeutic drugs bisphenol and <scene name='50/501401/Cv/5'>tamoxifen</scene> are nestled between 4 alpha helices in the ERR active site. | |||
* [[Estrogen sulfotransferase]] | |||
*[[Aromatase]] | |||
*[[Finasteride]] | |||
'''Estrone''' | |||
*[[Estrogen receptor 3D structures]] | |||
*[[Estrone sulfatase]] | |||
*[[Sulfatase 3D structures]] | |||
*[[ABC transporter 3D structures]] | |||
*[[ABCG2 multidrug transporter]] | |||
Substrates, such as estrone sulfate, <scene name='83/832932/Cavity_1_-_use2/4'>form hydrogen bonds and stacking interactions</scene> with residues from each subunit in Cavity 1 of ABCG2 multidrug transporter. | |||
*[[Hydroxysteroid dehydrogenase]] | |||
*[[Hydroxysteroid dehydrogenase 3D structures]] | |||
*[[Prostaglandin F synthase]] | |||
'''Estradiol''' | |||
*[[Lipids: structure and classification]] | |||
*[[Estrogen receptor]] | |||
*[[Ivan Koutsopatriy estrogen receptor]] | |||
*[[Estradiol 17-beta-dehydrogenase]] | |||
*[[Student Project 10 for UMass Chemistry 423 Spring 2015]] | |||
*[[Aromatase]] | |||
*[[Sulfotransferase]] | |||
*[[ATPase family AAA domain-containing protein 2]] | |||
*[[Hypoxia-Inducible Factors]] | |||
'''Estriol''' | |||
*[[Cytochrome P450 3D structures]]: [[1x8v]] - MtP450 CYP51 (mutant)+estriol<br /> | |||
'''Estetrol''' | |||
*[[3l03]] - Crystal Structure of human Estrogen Receptor alpha Ligand-Binding Domain in complex with a Glucocorticoid Receptor Interacting Protein 1 Nr Box II peptide and Estetrol (Estra-1,3,5(10)-triene-3,15 alpha,16alpha,17beta-tetrol) | |||
==[[Progestogens]]== | |||
'''Progesterone''' | |||
<scene name='89/895670/Cv/7'>Progesterone</scene> (P4) is an endogenous steroid and progestogen sex hormone involved in the menstrual cycle, pregnancy, and embryogenesis of humans and other species. | |||
*[[Progesterone receptor]]. <scene name='89/895670/Cv/8'>Human progesterone receptor ligand-binding domain bound with progesterone</scene> ([[1a28]]). Water molecules are shown as red spheres. | |||
*Progesterone is a negative allosteric modulator of [[nicotinic acetylcholine receptors]], and a potent antagonist of the [[mineralocorticoid receptor]]. | |||
*[[Hydroxysteroid dehydrogenase]], 20-α HSD is involved in the control of progesterone level in pregnancy of mice. 17-β HSD is involved in the conversion of androstenedione to testosterone. | |||
=Vitamin D derivatives; secosteroids (open-ring steroids)= | |||
<scene name='89/895670/Cv/9'>Vitamin D</scene>. | |||
<scene name='89/895670/Cv/10'>25-hydroxy-cholecalciferol (25-D3); 25-hydroxyvitamin D3</scene> ([[5ien]]) | |||
Calcitriol is the active form of vitamin D pro-hormone. | |||
*[[Vitamin D receptor]] (also called calcitriol receptor) | |||
<scene name='56/562378/Vit_d_receptor_3m7r/3'>Vitamin D receptor (VDR)</scene> is a transcription factor. Upon binding to vitamin D, VDR forms a heterodimer with retinoid-X receptor and binds to hormone response receptors on DNA causing gene expression. The <scene name='56/562378/Vit_d_receptor_ligand/1'>vitamin D hormone</scene> (green) binds to receptors in its target cells, controlling the synthesis of many different proteins involved in Ca transport and utilization. | |||
<scene name='51/517370/Cv/2'>Vitamin D hormone binding site</scene>. | |||
<scene name='51/517370/Cv/3'>Vitamin D hormone is located in deep pocket</scene>. VDR contains 2 domains: a <scene name='56/562378/Lbd/1'>ligand binding domain (LBD)</scene>, that binds to the hormone (grey) and <scene name='56/562378/Dbd/2'>DNA-binding domain (DBD)</scene> that binds to DNA (green and blue are 2 same VDR structures). It pairs up with a similar protein, 9-cis retinoic acid receptor (RXR), and together they bind to the DNA, activating synthesis in some cases and repressing it in others. When <scene name='56/562378/Serine_final/1'>serine</scene> is mutated it is replaced with a <scene name='56/562378/Glycine_final/1'>glycine</scene> which results in an inhibition of transcriptional activation. When transcription is inhibited it results in p53 accumulation, which activates and promotes p53 translocation into mitochondria leading to apoptosis. <scene name='56/562378/Serine_final/1'>Serine</scene> is replaced with <scene name='56/562378/Asparticacid_final/1'>aspartic acid</scene> when mutated creating a negative charge. The negative charge at the residue inhibits DNA binding which cause a downregulation of VDR activity. VDR needs DNA binding in order for it to be activated which is only possible with a serine residue. | |||
The vitamin D nuclear receptor is a ligand-dependent transcription factor that controls multiple biological responses such as cell proliferation, immune responses, and bone mineralization. Numerous 1 α,25(OH)(2)D(3) analogues, which exhibit low calcemic side effects and/or antitumoral properties, have been synthesized. It was shown that <scene name='56/562378/3a3z/1'>the synthetic analogue (20S,23S)-epoxymethano-1α,25-dihydroxyvitamin D(3) (2a)</scene> acts as a 1α,25(OH)(2)D(3) superagonist and exhibits both antiproliferative and prodifferentiating properties in vitro. Using this information and on the basis of the crystal structures of human VDR ligand binding domain (hVDR LBD) bound to 1α,25(OH)(2)D(3), 2α-methyl-1α,25(OH)(2)D(3), or 2a, a novel analogue, 2α-methyl-(20S,23S)-epoxymethano-1α,25-dihydroxyvitamin D(3) (4a) was designed, in order to increase its transactivation potency. | |||
'''Signaling Pathways:''' | '''Signaling Pathways:''' | ||
Revision as of 17:55, 15 December 2021
Under development!
Sphingosine-1-Phosphate Glucosylceramide Phosphatidylinositol bisphosphate (PIP2) Phosphatidylinositol 4,5-bisphosphate (PIP2) binds to and directly activates inwardly rectifying potassium channels. Inward rectifier KCh.
Retinal
Retinoic acid
Steroid Hormones and their receptors This large and diverse class of steroids are biosynthesized from isoprenoids and structurally resemble cholesterol. Mammalian steroid hormones can be grouped into five groups by the receptors to which they bind: glucocorticoids, mineralocorticoids, androgens, estrogens, and progestogens. Vitamin D derivatives are a sixth closely related hormone system with homologous receptors. They have some of the characteristics of true steroids as receptor ligands. For example, is an important estrogen steroid hormone in both women and men. It is a typical steroid with core four-ring system (ABCD), composed of 17 carbon atoms. Corticosteroids are a class of steroid hormones that are produced in the adrenal cortex of vertebrates, as well as the synthetic analogues of these hormones. Two main classes of corticosteroids, glucocorticoids and mineralocorticoids, are involved in a wide range of physiological processes. and its derivatives have some mineralocorticoid action in addition to the glucocorticoid effect.
(hydrocortisone) is a corticosteroid with both glucocorticoid and mineralocorticoid activity and effects. Glucocorticoids are corticosteroids that bind to the glucocorticoid receptor. is a glucocorticoid medication. It is the most potent glucocorticoid and it has not mineralocorticoid potency.
Mineralocorticoids are a class of corticosteroids. Mineralocorticoids are produced in the adrenal cortex and influence salt and water balances (electrolyte balance and fluid balance). The primary mineralocorticoid is .
Sex steroidsAndrogensAn androgen is any natural or synthetic steroid hormone that regulates the development and maintenance of male characteristics in vertebrates by binding to androgen receptors. The major androgen in males is . It is the primary sex hormone and anabolic steroid in males. It is a steroid from the androstane class. It exerts its action through binding to and activation of the androgen receptor.
EstrogensThere are three major endogenous estrogens that have estrogenic hormonal activity: estrone (E1), estradiol (E2), and estriol (E3). Estradiol, an estrane, is the most potent and prevalent. Another estrogen called estetrol (E4) is produced only during pregnancy. of estrogen receptor α complexed with raloxifene and a corepressor peptide (morph was taken from Gallery of Morphs of the Yale Morph Server). Structure of estradiol metal chelate and estrogen receptor complex: The basis for designing a new class of SERMs[23]. Selective estrogen receptor modulators, such as estradiol 17-derived metal complexes, have been synthesized as targeted probes for the diagnosis and treatment of breast cancer. The detailed 3D structure of bound with a novel at 2.6Å resolution was reported (2yat). The residues with EPTA-Eu. The hydrogen bonds are shown as white dashed lines. of this structure with the structure of native ligand 17β-estradiol (E2) in the complex of E2/ERα-LBD complex (1ere) reveals that the . The made by additional estrogen receptor residues (e.g. Glu419 of H7 and Glu339 of H3, this depends on subunit), may work together with the E2 17β hydroxyl-His524 hydrogen bond and tighten the neck of the LBP upon binding of the endogenous ligand E2. 4-Hydroxytamoxifen (OHT) is an other selective estrogen receptor modulator. of EPTA-Eu/ERα-LBD complex on OHT/ERα-LBD complex (3ert) shows that there is similar network of hydrogen bonds in both complexes, except for His524 which does not form hydrogen bond with OHT in the OHT/ERα-LBD complex. E2/ERα-LBD (1ere), OHT/ERα-LBD (3ert) and EPTA-Eu/ERα-LBD shows that they overlap well in the majority portions of the domain, but differ significantly in the region of the 'omega loop'. They display different synergistic reciprocating movements, depending on the specific nature of the ligand bound. The structure of estrogen receptor complexed with EPTA-Eu provides important information pertinent to the design of novel functional ER targeted probes for clinical applications. ER is a modular protein composed of a ligand binding domain, a DNA binding domain and a transactivation domain. ER is a DNA-binding transcription factor. . The DNA binding domain can be clearly observed in this scene; the highlighted yellow helix in close proximity to the DNA is part of the DNA binding domain. The blue beta sheet close to the yellow DNA binding alpha helix is also part of the DNA binding domain. The transactivation domain forms an alpha helix which is colored in purple. The transactivation domain activates RNA polymerase when the receptor binds to DNA. The ligand binding domain may be observed here with the following scene. . The ligand ferutinine (highlighted in pink) is bound by the ligand binding domain, composed of the blue colored alpha helices immediately surrounding the purple ligand. Another view of the ligand binding domain is shown here, with estradiol bound. . ER is functional as a ligand-dependent transcription factor. ER responds to both agonist and antagonist ligands and can associate with the nuclear matrix. Differences in the structure of the receptor are observed depending on what ligand ER has bound (if any). Through comparisons of ER bound to agonist and antagonist ligands, some structural components may be highlighted. The specific conformation of this tight loop of alpha helices and beta sheets around the ligand shows a complex capable of activating ER's transcription loci. This complex allows for the activation signal that will stimulate normal growth. Normal growth is stimulated when an agonist bound ER binds DNA. This occurs with the assistance of chaperon proteins. These chaperons are capable of recognizing estrogen receptor ligand complexes. When ER has bound a ligand chaperons facilitate the trans-location of the complex to the nucleus. Eventually the chaperon ligand ER complex will reach specific euchromatin, at which point the chaperons facilitate the ligand ER complex to changes conformation. This conformation will facilitate the estrogen receptor to bind the DNA major groove at specific palindromic sequences. Estradiol is a normal ligand for ER and allows for binding in the major groove of DNA. If the ligand is an antagonist the transcription factor function of estrogen receptor becomes hindered. The conformation of ER bound to the partial agonist genistein has a loop which is not as tight around the ligand as those found on ER with a complete agonist ligand. The ligands themselves take up different amounts of space and have varying interactions within ER. This slight difference effects the ability of the chaperon to be able to bind the receptor ligand complex to the major groove of DNA. There is a noticeable difference in the size of the pure agonist vs partial agonist scenes. Specifically, look at the width of the agonist compared to the partial agonist. Similar differences may be observed between ER which has bound the partial agonist and complete antagonist ligands. The most drastic difference is noticeable between agonist and antagonist ligands. Compare the agonist scene to the . Special attention should be given to the bottom right alpha helices and beta sheets that are pushed out more in the antagonist compared to the agonist bound ER.
(ER-β) is 1 of the 2 isoforms of the estrogen receptor, a ligand-activated transcription factor which regulates the biological effects of the steroid hormone 17 β-estradiol, or estrogen, in both males and females. The complex is a hetero-tetrameric assembly consisting of 4 molecules and a ligand: 2 copies of , 2 copies of , and the ligand, . Once the ligand is bound, the complex recruits the steroid receptor coactivators, which recruit other proteins to form the transcriptional complex for initiation of transcription. This activates expression of reporter genes containing estrogen response elements. Genistein is a phytoestrogen with structural similarity to estrogen which competes for estrogen receptors. Although estrogen receptor β is widely expressed, it is not the primary estrogen receptor in most tissues. As a result, it has become a target for drug delivery, especially since it is 40x more selective for genistein than the α isoform. This enhanced selectivity may be caused by differences in residues between the 2 isoforms, allowing ER-β to accommodate more polar substituents in its binding pocket. ER-β differs greatly from ER-α at the N-terminal domains, which can be seen located at opposite ends from the C termini in this . The protein is composed of 3 sections: a modulating N-terminal domain, a DNA-binding domain and a C-terminal ligand-binding domain.
Each ERβ contains several domains with specific functions: an N-terminal domain (NTD), a DNA-binding domain (DBD), a flexible hinge region and a C-terminal Ligand-binding domain (LBD). The complex overall is about . The is the 1st activation function (AF-1) domain that consists mostly of random coils and a small portion of helices (red) and sheets (green); it is a . This lack of structure allows the region to recruit and bond many different interaction partners. This region also has the capacity to transactivate transcription without binding estrogen. The binds estrogen response elements (ERE) of target genes and recruits coactivator proteins responsible for the transcription of these genes. The ERE consist of a palindromic inverted repeat 5'GGTCAnnnTGACC-3' of target genes. The DBD is a highly . It is composed of 2 C4-type Zn fingers each containing residues coordinating to a Zn atom. The hinge region connects the DBD and LBD. binds estrogen, coregulatory proteins, corepressors and coactivators. Genistein is not generated by the endocrine system that binds ERβ like estrogen; both ligands are completely buried within the (Hydrophobic, Polar) of the ERβ complex. Binding at the LBD activates transcription mediated by the DBD. This domain is entirely helical; the LBD interacts with genistein through helices. The conformationally dynamic portion of this region gives rise to ERβ’s ligand-dependent transcriptional activation (AF-2) function. A key element of AF-2 is helix 12 (H12), which acts as a conformational switch; different receptor ligands influence the orientation of H12. Agonist ligands like genistein position H12 across the ligand-binding pocket of the LBD, which provides a coactivator docking surface. Geinstein binding allows the helices of AF-2 to form a shallow hydrophobic binding site for leucine-rich motifs of coactivators to bind. This conformation provides optimal interaction with coactivators and transcription is activated. Genistein's bicyclic form allows it to hydrogen bond on opposite sides with the hydroxyls of the histidine groups on the receptor. binding to the receptor causes a conformational change and activates the receptor resulting in up-regulation for coactivators. Down-regulation will occur in the presence of corepressor as they bind to repressors and indirectly regulate gene expression. In order for the estrogen receptor β genistein to bind to a receptor and activate it there must be stabilization by a coactivator. The coactivator increases the gene expression and with this increase allows it to bind to an activator group consisting of a DNA binding domain. The estrogen receptor is found to be comprised of a dimer attached to a ligand and coactivator peptide which helps to stabilize the structure of each monomer. The conformational state of helix-12 can be modified by the binding of the coactivator. This depicts the hydrophobic and hydrophilic residues of the estrogen receptor. The hydrophobic regions are primarily on the inside of the protein surrounding genistein (red). Having the hydrophobic residues surrounding the binding pocket will stabilize the structure. The structure of this pocket is tertiary and do to the hydrophobic interactions inside the pocket and hydrophilic interactions on the outside help to stabilize this tertiary structure. The is hydrophobic which means that an increase in lipophilicity would increase the affinity for ligands which in this case is genistein. The genistein structure has 3 hydroxyl groups, an ether and an ester. These 3 functional groups are polar and have many possibilities for hydrogen bonding. The His475 and Met336 residues in the binding pocket are capable of forming hydrogen bonds with genistein do to the many hydrogen bond forming functional groups. These residues are different from the residues found in ERα and so the selectivity of genistein is much greater for ERβ. Upon visualizing the estrogen receptor in an , the structure can be classified as parallel or anti-parallel. Here is the zoomed .
to human estrogen-related receptor γ. The chemotherapeutic drugs bisphenol and are nestled between 4 alpha helices in the ERR active site. Estrone
Substrates, such as estrone sulfate, with residues from each subunit in Cavity 1 of ABCG2 multidrug transporter. Estradiol
Estriol
Estetrol
ProgestogensProgesterone (P4) is an endogenous steroid and progestogen sex hormone involved in the menstrual cycle, pregnancy, and embryogenesis of humans and other species.
Vitamin D derivatives; secosteroids (open-ring steroids). (5ien) Calcitriol is the active form of vitamin D pro-hormone.
is a transcription factor. Upon binding to vitamin D, VDR forms a heterodimer with retinoid-X receptor and binds to hormone response receptors on DNA causing gene expression. The (green) binds to receptors in its target cells, controlling the synthesis of many different proteins involved in Ca transport and utilization. . . VDR contains 2 domains: a , that binds to the hormone (grey) and that binds to DNA (green and blue are 2 same VDR structures). It pairs up with a similar protein, 9-cis retinoic acid receptor (RXR), and together they bind to the DNA, activating synthesis in some cases and repressing it in others. When is mutated it is replaced with a which results in an inhibition of transcriptional activation. When transcription is inhibited it results in p53 accumulation, which activates and promotes p53 translocation into mitochondria leading to apoptosis. is replaced with when mutated creating a negative charge. The negative charge at the residue inhibits DNA binding which cause a downregulation of VDR activity. VDR needs DNA binding in order for it to be activated which is only possible with a serine residue. The vitamin D nuclear receptor is a ligand-dependent transcription factor that controls multiple biological responses such as cell proliferation, immune responses, and bone mineralization. Numerous 1 α,25(OH)(2)D(3) analogues, which exhibit low calcemic side effects and/or antitumoral properties, have been synthesized. It was shown that acts as a 1α,25(OH)(2)D(3) superagonist and exhibits both antiproliferative and prodifferentiating properties in vitro. Using this information and on the basis of the crystal structures of human VDR ligand binding domain (hVDR LBD) bound to 1α,25(OH)(2)D(3), 2α-methyl-1α,25(OH)(2)D(3), or 2a, a novel analogue, 2α-methyl-(20S,23S)-epoxymethano-1α,25-dihydroxyvitamin D(3) (4a) was designed, in order to increase its transactivation potency. Signaling Pathways: ABA Signaling Pathway
Tyrosine kinase
Protein kinase C
MAPK
CAMP-dependent protein kinase
Transient receptor potential channels
Light is detected by rhodopsin in rod and cone cells.
Protein phosphatases: Second messengers
CAMP-dependent protein kinase Receptors that activate this pathway (Phospholipase C) are mainly G protein-coupled receptors coupled to the Gαq subunit, including:
Paracrine signaling: fibroblast growth factor (FGF) family, Hedgehog family, Wnt family, and TGF-β superfamily Fibroblast growth factor and Fibroblast growth factor receptor (FGFR). FGFR belongs to Receptor tyrosine kinases, class V. Sonic Hedgehog
Ca2+ signalling processes
H+/K+-ATPase signal pathway (acetylcholine, histamine, and gastrin) activates the pump in order to move the vesicles toward the lumen. Proton pump Signal transducing adaptor proteins (STAPs)
GTPase The Mitogen-activated protein kinase cascade MAPKs are involved in directing cellular responses to a diverse array of stimuli, such as mitogens, osmotic stress, heat shock and proinflammatory cytokines. They regulate cell functions including proliferation, gene expression, differentiation, mitosis, cell survival, and apoptosis.
Inflammatory response
Allostery ATPase
|
| |||||||||||||||||||
ReferencesReferences
- ↑ Chatterjee S. Neutral sphingomyelinase: past, present and future. Chem Phys Lipids. 1999 Nov;102(1-2):79-96. PMID:11001563
- ↑ Barna TM, Khan H, Bruce NC, Barsukov I, Scrutton NS, Moody PC. Crystal structure of pentaerythritol tetranitrate reductase: "flipped" binding geometries for steroid substrates in different redox states of the enzyme. J Mol Biol. 2001 Jul 6;310(2):433-47. PMID:11428899 doi:10.1006/jmbi.2001.4779
- ↑ Tuteja G, Kaestner KH. SnapShot: forkhead transcription factors I. Cell. 2007 Sep 21;130(6):1160. PMID:17889656 doi:http://dx.doi.org/10.1016/j.cell.2007.09.005
- ↑ Kaiser G, Gerst F, Michael D, Berchtold S, Friedrich B, Strutz-Seebohm N, Lang F, Haring HU, Ullrich S. Regulation of forkhead box O1 (FOXO1) by protein kinase B and glucocorticoids: different mechanisms of induction of beta cell death in vitro. Diabetologia. 2013 Jul;56(7):1587-95. doi: 10.1007/s00125-013-2863-7. Epub 2013, Feb 23. PMID:23435785 doi:http://dx.doi.org/10.1007/s00125-013-2863-7
- ↑ Horwitz KB, Jackson TA, Bain DL, Richer JK, Takimoto GS, Tung L. Nuclear receptor coactivators and corepressors. Mol Endocrinol. 1996 Oct;10(10):1167-77. PMID:9121485 doi:http://dx.doi.org/10.1210/mend.10.10.9121485
- ↑ Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000 Oct;267(20):6102-9. PMID:11012661
- ↑ Prasad R, Chan LF, Hughes CR, Kaski JP, Kowalczyk JC, Savage MO, Peters CJ, Nathwani N, Clark AJ, Storr HL, Metherell LA. Thioredoxin Reductase 2 (TXNRD2) mutation associated with familial glucocorticoid deficiency (FGD). J Clin Endocrinol Metab. 2014 Aug;99(8):E1556-63. doi: 10.1210/jc.2013-3844. Epub, 2014 Mar 6. PMID:24601690 doi:http://dx.doi.org/10.1210/jc.2013-3844
- ↑ Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000 Oct;267(20):6102-9. PMID:11012661
- ↑ Fritz-Wolf K, Kehr S, Stumpf M, Rahlfs S, Becker K. Crystal structure of the human thioredoxin reductase-thioredoxin complex. Nat Commun. 2011 Jul 12;2:383. doi: 10.1038/ncomms1382. PMID:21750537 doi:10.1038/ncomms1382
- ↑ Murakami M, Nakatani Y, Tanioka T, Kudo I. Prostaglandin E synthase. Prostaglandins Other Lipid Mediat. 2002 Aug;68-69:383-99. PMID:12432931
- ↑ Kudo I, Murakami M. Prostaglandin E synthase, a terminal enzyme for prostaglandin E2 biosynthesis. J Biochem Mol Biol. 2005 Nov 30;38(6):633-8. PMID:16336776
- ↑ Luz JG, Antonysamy S, Kuklish SL, Condon B, Lee MR, Allison D, Yu XP, Chandrasekhar S, Backer R, Zhang A, Russell M, Chang SS, Harvey A, Sloan AV, Fisher MJ. Crystal Structures of mPGES-1 Inhibitor Complexes Form a Basis for the Rational Design of Potent Analgesic and Anti-Inflammatory Therapeutics. J Med Chem. 2015 May 20. PMID:25961169 doi:http://dx.doi.org/10.1021/acs.jmedchem.5b00330
- ↑ Frey FJ, Odermatt A, Frey BM. Glucocorticoid-mediated mineralocorticoid receptor activation and hypertension. Curr Opin Nephrol Hypertens. 2004 Jul;13(4):451-8. PMID:15199296
- ↑ Pujo L, Fagart J, Gary F, Papadimitriou DT, Claes A, Jeunemaitre X, Zennaro MC. Mineralocorticoid receptor mutations are the principal cause of renal type 1 pseudohypoaldosteronism. Hum Mutat. 2007 Jan;28(1):33-40. PMID:16972228 doi:10.1002/humu.20371
- ↑ Geller DS, Farhi A, Pinkerton N, Fradley M, Moritz M, Spitzer A, Meinke G, Tsai FT, Sigler PB, Lifton RP. Activating mineralocorticoid receptor mutation in hypertension exacerbated by pregnancy. Science. 2000 Jul 7;289(5476):119-23. PMID:10884226
- ↑ Lother A, Bergemann S, Kowalski J, Huck M, Gilsbach R, Bode C, Hein L. Inhibition of the cardiac myocyte mineralocorticoid receptor ameliorates doxorubicin-induced cardiotoxicity. Cardiovasc Res. 2018 Feb 1;114(2):282-290. doi: 10.1093/cvr/cvx078. PMID:28430882 doi:http://dx.doi.org/10.1093/cvr/cvx078
- ↑ Caprio M, Feve B, Claes A, Viengchareun S, Lombes M, Zennaro MC. Pivotal role of the mineralocorticoid receptor in corticosteroid-induced adipogenesis. FASEB J. 2007 Jul;21(9):2185-94. doi: 10.1096/fj.06-7970com. Epub 2007 Mar 23. PMID:17384139 doi:http://dx.doi.org/10.1096/fj.06-7970com
- ↑ Bledsoe RK, Madauss KP, Holt JA, Apolito CJ, Lambert MH, Pearce KH, Stanley TB, Stewart EL, Trump RP, Willson TM, Williams SP. A ligand-mediated hydrogen bond network required for the activation of the mineralocorticoid receptor. J Biol Chem. 2005 Sep 2;280(35):31283-93. Epub 2005 Jun 20. PMID:15967794 doi:http://dx.doi.org/10.1074/jbc.M504098200
- ↑ Bohl CE, Wu Z, Chen J, Mohler ML, Yang J, Hwang DJ, Mustafa S, Miller DD, Bell CE, Dalton JT. Effect of B-ring substitution pattern on binding mode of propionamide selective androgen receptor modulators. Bioorg Med Chem Lett. 2008 Oct 15;18(20):5567-70. Epub 2008 Sep 5. PMID:18805694 doi:10.1016/j.bmcl.2008.09.002
- ↑ Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993 Mar;13(3):1392-407. PMID:8441385
- ↑ Kondo N, Katsuno M, Adachi H, Minamiyama M, Doi H, Matsumoto S, Miyazaki Y, Iida M, Tohnai G, Nakatsuji H, Ishigaki S, Fujioka Y, Watanabe H, Tanaka F, Nakai A, Sobue G. Heat shock factor-1 influences pathological lesion distribution of polyglutamine-induced neurodegeneration. Nat Commun. 2013;4:1405. doi: 10.1038/ncomms2417. PMID:23360996 doi:http://dx.doi.org/10.1038/ncomms2417
- ↑ Ghosh, D., Griswold, J., Erman, M., Pangborn, W. " X-ray Structure of Human Aromatase Reveals An Androgen-Specific Active Site" Journal of Steroid Biochemistry and Molecular Biology. [Online] 2010,Vol. 118, Issue 4-5, p197-202[1]
- ↑ Li MJ, Greenblatt HM, Dym O, Albeck S, Pais A, Gunanathan C, Milstein D, Degani H, Sussman JL. Structure of estradiol metal chelate and estrogen receptor complex: The basis for designing a new class of selective estrogen receptor modulators. J Med Chem. 2011 Apr 7. PMID:21473635 doi:10.1021/jm200192y