Nos1
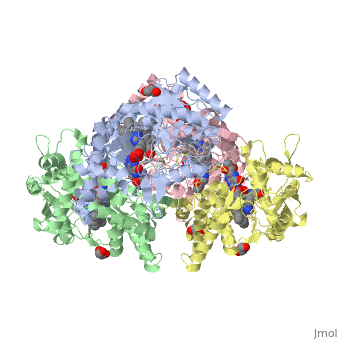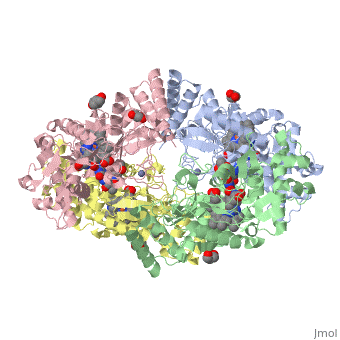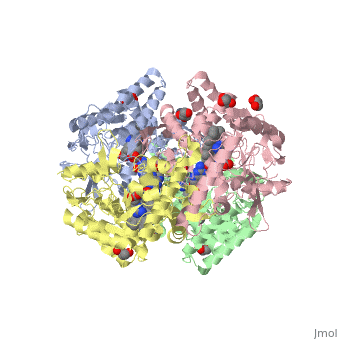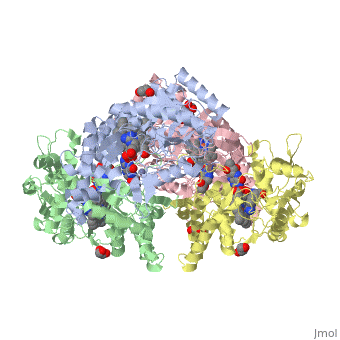
LocationIn humans, three nitric oxide synthase isoforms are expressed which include NOS1 (neuronal NOS or nNOS), NOS2 (inducible NOS or iNOS), and NOS3 (endothelial NOS or eNOS) (see Nitric Oxide Synthase).[1] NOS1 is located on chromosome 12[2] and is expressed in all tissues but has displayed high expression in skeletal muscle as well as in brain, testicular, lung, and kidney tissues.[3] Moderate expression has also been observed in heart, adrenal gland, and retinal tissues.[3] The top ten out of 448 tissues with the highest expression of NOS1 are shown here, all of which are either brain tissue, skeletal muscles, or eye muscles.[4] Figure 1. Microarray expression data for human NOS1 across 448 tissues. Nitric oxide synthases consume L-arginine and molecular oxygen to form the free radical nitric oxide (NO), which acts as a signaling molecule in a wide range of molecular and biological processes.[5] NOS1 is suggested to be a membrane-bound enzyme as its activity was observed to be localized to plasma membranes. NOS1 activity is directly associated with neuronal dendritic spines and to sarcolemma of skeletal and cardiac muscle cells according to multiple localization studies.[6][7][8] The reaction catalyzed by nitric oxide synthase is shown below.[2] 2 L-arginine + 3 NADPH + 4 O(2) = 2 L- citrulline + 2 nitric oxide + 3 NADP(+) + 4 H(2)O FunctionNOS1 is a major component of the production of nitric oxide. Nitric oxide targets nitric oxide-specific receptors such as G-cyclase. Nitric oxide binds to the receptors to act as a major signaling molecule. NOS1 is an important component of synaptogenesis, long-term potentiation, neurotransmitter release and synaptic plasticity.[9] Synaptogenesis, long-term potentiation, neurotransmitter release and synaptic plasticity occur when NOS1 is coupled with N-methyl-D-Aspartate (NMDA) receptors in the post-synapse of cells. NMDA receptor binds to PSD95, which then binds to NOS1. After the complex is formed, NMDA receptor mediated CA2+ influx now regulates the amount of nitric oxide that is produced by NOS1.[10] NOS1 also contributes in limiting oxidative stress in the myocardium, limits systolic and diastolic dysfunction and prevents a failing heart. NOS1 regulates the reuptake of Ca2+ in sarcoplamsic reticulum (SR). NOS1 is shown into the human coronary artery smooth cells and maintains of basal blood flow. NOS1 also controls the parasympathetic and sympathetic regulation of the heart.[11] NOS1 is important in all functions that help regulate the cardiac muscles.[11] The ability of nitric oxide to self regulate enables NOS1 to control all these major functions. In myocyte relaxation, the nitric oxide produced by NOS1 facilitates SERCA to increase intracellular CA2+ because of the increase in PKA dependent PLN phosphorylation. DiseaseNitric oxide is produced from one of three synthases present within the body: neuronal nitric oxide synthase (NOS1), inducible nitric oxide synthase (NOS2), and endothelial nitric oxide synthase (NOS3).[12] Specifically, NOS1 has nine different first exons that lead to multiple NOS1 transcripts with different 5’-untranslated regions.[13] An advantage of having multiple first exons that can be alternatively spliced and expressed is that NOS1 can be specific and specifically regulated for different tissues.[13] A disadvantage to having multiple first exons is that there is a higher possibility of mutations, which would affect NO production and thus have an effect on the second messenger cyclic guanine monophosphate (cGMP) production.[12] Furthermore, because NO is an oxyradical, overproduction caused by a mutation can lead to neural tissue damage.[12] Numerous pathologies may arise from neural tissue damage, and one study suggested overproduction of NO that leads to such damage is possibly an influential factor in developing schizophrenia.[12] Single nucleotide polymorphisms (SNPs) and various lengths of tandem repeats within NOS1 have been linked in other disorders of the brain such as Alzheimer’s and Parkinson’s diseases.[13][14] Out of three identified SNPs occurring in alternative exon 1c, only the SNP G-84A has a functional effect that decreases transcription levels.[13] However, various lengths of tandem repeats present in the alternative exon have been shown to be a potential factor in both Alzheimer’s and Parkinson’s diseases.[13][14] Shorter tandem TG repeats are possibly associated with the development of Alzheimer’s disease, and longer tandem TG repeats are possibly associated with the development of Parkinson’s disease.[14] Although schizophrenia, Alzheimer’s, and Parkinson’s diseases have genetic influences, mutations in NOS1 can be a risk indicator for developing these diseases.[12] [13][14] Structural highlightsNeuronal nitric oxide synthase exists as a homodimer. This homodimer consists of two regions. One region is an N-terminal oxygenase domain and the other is a C-terminal reductase. The N-terminal oxygenase domain is an extended beta sheet cage with binding sites for heme. The N-terminal catalytic domain contains a , a nearby cofactor site for tetrahydrobiopterin () and a C-terminal reductase domain that consists of FMN, FAD, and NADPH binding sites.[15] The structure of the human nNOS heme domain contains cysteine and tryptophan. The NADPH provides electrons for catalysis. These electrons are first passed to FAD and FMN and then the heme. This electron flow is monitored by the binding of CaM and Calcium2+ in the linker region between the two heterodimer domains. The amino acid composition of asparagine and methionine in rat neuronal NOS are conserved in human nNOS. This amino acid composition influences the binding of inhibitors and makes nNOS inhibitors highly selective. The structure of the human nNOS heme domain contains cysteine and tryptophan. The cysteine subunits of one region pack closely against the backbone of histidine in subunits of another region. Crystal packing interactions most likely contribute to the fully ordered N-termini in the NOS structure. There is a flexible surface loop downstream from the binding site. This loop in human nNOS lacks any secondary structure and appears as a random coil. This reflects the flexible nature of nNOS because it can easily adapt to local environment which involves crystal packing. This loop is found near the CaM binding site and can therefore most likely interact with it and affect electron flow. [16]
|
| ||||||||||
ReferencesReferences
- ↑ Stuehr DJ. (May 1999) "Mammalian nitric oxide synthases". Biochim. Biophys. Acta 1411 (2–3): 217–30. doi:10.1016/S0005-2728(99)00016-X. PMID 10320659.
- ↑ 2.0 2.1 Knowles RG, Moncada S. (March 1994) "Nitric oxide synthases in mammals". Biochem. J. 298 (2): 249–58. PMC 1137932. PMID 7510950.
- ↑ 3.0 3.1 Ward ME, Toporsian M, Scott JA, et al. (2005) Hypoxia induces a functionally significant and translationally efficient neuronal NO synthase mRNA variant. Journal of Clinical Investigation. 115(11):3128-3139. doi:10.1172/JCI20806.
- ↑ Genevisible (ND) 'Expression of P29475' [online] available at https://genevisible.com/tissues/HS/UniProt/P29475
- ↑ Hou, YC; Janczuk, A; Wang, PG. (1999) "Current trends in the development of nitric oxide donors". Current pharmaceutical design 5 (6): 417–41.
- ↑ M Hecker, A Mulsch, R Busse. (1994) Subcellular localization and characterization of neuronal nitric oxide synthase. J. Neurochem., 62, pp. 1524–1529
- ↑ Beigi F, Oskouei BN, Zheng M, Cooke CA, Lamirault G, Hare JM. (2009) Cardiac nitric oxide synthase-1 localization within the cardiomyocyte is accompanied by the adaptor protein, CAPON. Nitric Oxide. 21:226–233. doi: 10.1016/j.niox.2009.09.005.
- ↑ Barouch LA, Harrison RW, Skaf MW, Rosas GO, Cappola TP, Kobeissi ZA, et al. (2002) Nitric oxide regulates the heart by spatial confinement of nitric oxide synthase isoforms. Nature. 416:337–9. doi: 10.1038/416337a
- ↑ Juliane Kopf, Martin Schecklmann, Tim Hahn, Thomas Dresler, Alica C. Dieler, Martin J. Herrmann, Andreas J. Fallgatter, Andreas Reif. (2011) NOS1 ex1f-VNTR polymorphism influences prefrontal brain oxygenation during a working memory task, NeuroImage, Volume 57, (Issue 4),1617-1623, Ihttp://dx.doi.org/10.1016/
- ↑ Freudenberg, F., Alttoa, A. & Reif, A. (2015) Neuronal nitric oxide synthase (NOS1) and its adaptor (NOS1AP) act as a genetic risk factors for psychiatric. Genes Brain Behav 14, 47–64.
- ↑ 11.0 11.1 Zhang, Y. H., Jin, C. Z., Jang, J. H., & Wang, Y. (2014). Molecular mechanisms of neuronal nitric oxide synthase in cardiac function and pathophysiology. The Journal of Physiology, 592(Pt 15), 3189–3200. http://doi.org/10.1113/jphysiol.2013.270306]
- ↑ 12.0 12.1 12.2 12.3 12.4 Shinkai, T., Ohmori, O., Hori, H., and Nakamura, J. (2002) Allelic association of the neuronal nitric oxide synthase (NOS1) gene with schizophrenia. Molecular Psychiatry. 7, 560-563. doi:10.1038/sj.mp.4001041
- ↑ 13.0 13.1 13.2 13.3 13.4 13.5 Galimberti, D., Scarpini, E., Venturelli, E., Strobel, A., Herterich, S., Fenogolio, C., Guidi, I., Scalabrini, D., Cortini, F., Bresolin, N., Lesch, K., and Reif, A. (2008) Association of a NOS1 promoter repeat with Alzheimer’s disease. Neurobiology of Aging. 29, 1359-1365. doi:10.1016/j.neurobiolaging.2007.03.003
- ↑ 14.0 14.1 14.2 14.3 Rife, T., Rasoul, B., Pullen, N., Mitchell, D., Grathwol, K., and Kurth, J. (2009) The effect of a promoter polymorphism on transcription of nitric oxide synthase 1 and its relevance to Parkinson’s disease. Journal of Neuroscience Research. 87, 2319-2325. doi:10.1005/jnr.22045
- ↑ Li H, Jamal J, Plaza C, et al. Structures of human constitutive nitric oxide synthases. Acta Crystallographica Section D: Biological Crystallography. 2014;70(Pt 10):2667-2674.
- ↑ Li H, Jamal J, Plaza C, et al. Structures of human constitutive nitric oxide synthases. Acta Crystallographica Section D: Biological Crystallography. 2014;70(Pt 10):2667-2674.