4ivt: Difference between revisions
No edit summary |
No edit summary |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
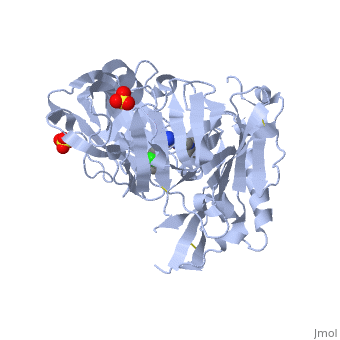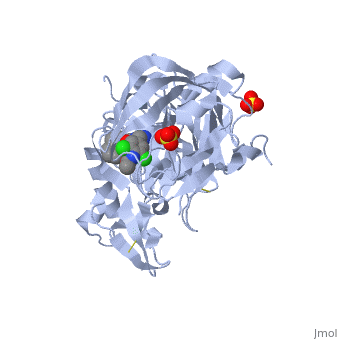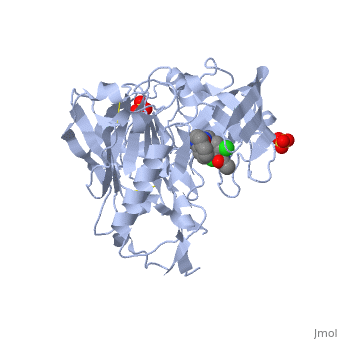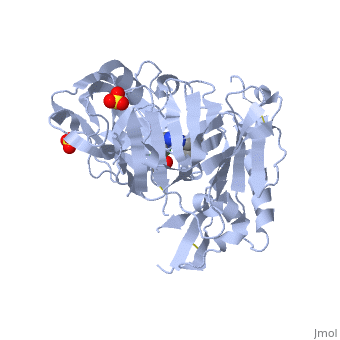
==Crystal structure of BACE1 with its inhibitor== | |||
<StructureSection load='4ivt' size='340' side='right'caption='[[4ivt]], [[Resolution|resolution]] 1.60Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[4ivt]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4IVT OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=4IVT FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.6Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene>, <scene name='pdbligand=VTI:N-{N-[4-(ACETYLAMINO)-3,5-DICHLOROBENZYL]CARBAMIMIDOYL}-2-(1H-INDOL-1-YL)ACETAMIDE'>VTI</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=4ivt FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4ivt OCA], [https://pdbe.org/4ivt PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=4ivt RCSB], [https://www.ebi.ac.uk/pdbsum/4ivt PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=4ivt ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/BACE1_HUMAN BACE1_HUMAN] Responsible for the proteolytic processing of the amyloid precursor protein (APP). Cleaves at the N-terminus of the A-beta peptide sequence, between residues 671 and 672 of APP, leads to the generation and extracellular release of beta-cleaved soluble APP, and a corresponding cell-associated C-terminal fragment which is later released by gamma-secretase.<ref>PMID:10677483</ref> <ref>PMID:20354142</ref> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Proteolytic cleavage of amyloid precursor protein by beta-secretase (BACE1) is a key step in generating the N-terminal of beta-amyloid (Abeta), which further forms into amyloid plaques that are considered as the hallmark of Alzheimer's disease. Inhibitors of BACE1 can reduce the levels of Abeta and thus have a therapeutic potential for treating the disease. We report here the identification of a series of small molecules bearing an indole acylguanidine core structure as potent BACE1 inhibitors. The initial weak fragment was discovered by virtual screening, and followed with a hit-to-lead optimization. With the aid of co-crystal structures of two discovered inhibitors (compounds 19 and 25) with BACE1, we explored the SAR around the indole and aryl groups, and obtained several BACE1 inhibitors about 1,000-fold more potent than the initial fragment hit. Accompanying the lead optimization, a previously under-explored sub-site opposite the flap loop was redefined as a potential binding site for later BACE1 inhibitor design. | |||
Virtual screening and structure-based discovery of indole acylguanidines as potent beta-secretase (BACE1) inhibitors.,Zou Y, Li L, Chen W, Chen T, Ma L, Wang X, Xiong B, Xu Y, Shen J Molecules. 2013 May 16;18(5):5706-22. doi: 10.3390/molecules18055706. PMID:23681056<ref>PMID:23681056</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 4ivt" style="background-color:#fffaf0;"></div> | |||
==See Also== | |||
*[[Beta secretase 3D structures|Beta secretase 3D structures]] | |||
*[[Beta-secretase 1|Beta-secretase 1]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Homo sapiens]] | |||
[[Category: Large Structures]] | |||
[[Category: Chen TT]] | |||
[[Category: Chen WY]] | |||
[[Category: Li L]] | |||
[[Category: Xu YC]] | |||
Latest revision as of 17:17, 8 November 2023
Crystal structure of BACE1 with its inhibitorCrystal structure of BACE1 with its inhibitor
Structural highlights
FunctionBACE1_HUMAN Responsible for the proteolytic processing of the amyloid precursor protein (APP). Cleaves at the N-terminus of the A-beta peptide sequence, between residues 671 and 672 of APP, leads to the generation and extracellular release of beta-cleaved soluble APP, and a corresponding cell-associated C-terminal fragment which is later released by gamma-secretase.[1] [2] Publication Abstract from PubMedProteolytic cleavage of amyloid precursor protein by beta-secretase (BACE1) is a key step in generating the N-terminal of beta-amyloid (Abeta), which further forms into amyloid plaques that are considered as the hallmark of Alzheimer's disease. Inhibitors of BACE1 can reduce the levels of Abeta and thus have a therapeutic potential for treating the disease. We report here the identification of a series of small molecules bearing an indole acylguanidine core structure as potent BACE1 inhibitors. The initial weak fragment was discovered by virtual screening, and followed with a hit-to-lead optimization. With the aid of co-crystal structures of two discovered inhibitors (compounds 19 and 25) with BACE1, we explored the SAR around the indole and aryl groups, and obtained several BACE1 inhibitors about 1,000-fold more potent than the initial fragment hit. Accompanying the lead optimization, a previously under-explored sub-site opposite the flap loop was redefined as a potential binding site for later BACE1 inhibitor design. Virtual screening and structure-based discovery of indole acylguanidines as potent beta-secretase (BACE1) inhibitors.,Zou Y, Li L, Chen W, Chen T, Ma L, Wang X, Xiong B, Xu Y, Shen J Molecules. 2013 May 16;18(5):5706-22. doi: 10.3390/molecules18055706. PMID:23681056[3] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||