2pbd: Difference between revisions
No edit summary |
No edit summary |
||
| (12 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
< | ==Ternary complex of profilin-actin with the poly-PRO-GAB domain of VASP*== | ||
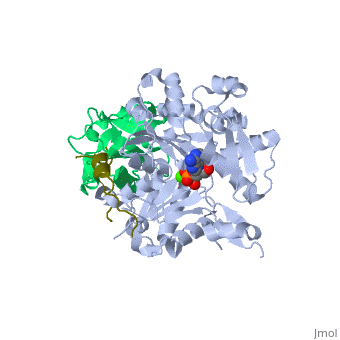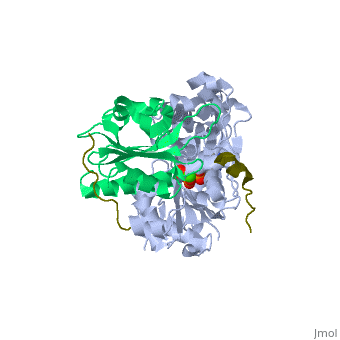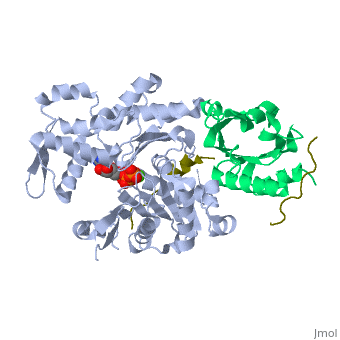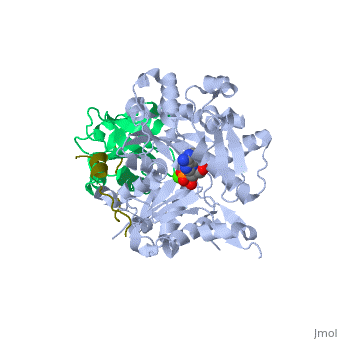
<StructureSection load='2pbd' size='340' side='right'caption='[[2pbd]], [[Resolution|resolution]] 1.50Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[2pbd]] is a 3 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens] and [https://en.wikipedia.org/wiki/Oryctolagus_cuniculus Oryctolagus cuniculus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=2PBD OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=2PBD FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.501Å</td></tr> | |||
- | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=ATP:ADENOSINE-5-TRIPHOSPHATE'>ATP</scene>, <scene name='pdbligand=CA:CALCIUM+ION'>CA</scene>, <scene name='pdbligand=HIC:4-METHYL-HISTIDINE'>HIC</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=2pbd FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=2pbd OCA], [https://pdbe.org/2pbd PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=2pbd RCSB], [https://www.ebi.ac.uk/pdbsum/2pbd PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=2pbd ProSAT]</span></td></tr> | |||
</table> | |||
== Disease == | |||
[https://www.uniprot.org/uniprot/PROF1_HUMAN PROF1_HUMAN] Defects in PFN1 are the cause of amyotrophic lateral sclerosis 18 (ALS18) [MIM:[https://omim.org/entry/614808 614808]. A neurodegenerative disorder affecting upper motor neurons in the brain and lower motor neurons in the brain stem and spinal cord, resulting in fatal paralysis. Sensory abnormalities are absent. The pathologic hallmarks of the disease include pallor of the corticospinal tract due to loss of motor neurons, presence of ubiquitin-positive inclusions within surviving motor neurons, and deposition of pathologic aggregates. The etiology of amyotrophic lateral sclerosis is likely to be multifactorial, involving both genetic and environmental factors. The disease is inherited in 5-10% of the cases.<ref>PMID:22801503</ref> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/PROF1_HUMAN PROF1_HUMAN] Binds to actin and affects the structure of the cytoskeleton. At high concentrations, profilin prevents the polymerization of actin, whereas it enhances it at low concentrations. By binding to PIP2, it inhibits the formation of IP3 and DG. Inhibits androgen receptor (AR) and HTT aggregation and binding of G-actin is essential for its inhibition of AR.<ref>PMID:18573880</ref> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/pb/2pbd_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=2pbd ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Cells sustain high rates of actin filament elongation by maintaining a large pool of actin monomers above the critical concentration for polymerization. Profilin-actin complexes constitute the largest fraction of polymerization-competent actin monomers. Filament elongation factors such as Ena/VASP and formin catalyze the transition of profilin-actin from the cellular pool onto the barbed end of growing filaments. The molecular bases of this process are poorly understood. Here we present structural and energetic evidence for two consecutive steps of the elongation mechanism: the recruitment of profilin-actin by the last poly-Pro segment of vasodilator-stimulated phosphoprotein (VASP) and the binding of profilin-actin simultaneously to this poly-Pro and to the G-actin-binding (GAB) domain of VASP. The actin monomer bound at the GAB domain is proposed to be in position to join the barbed end of the growing filament concurrently with the release of profilin. | |||
Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP.,Ferron F, Rebowski G, Lee SH, Dominguez R EMBO J. 2007 Oct 31;26(21):4597-606. Epub 2007 Oct 4. PMID:17914456<ref>PMID:17914456</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 2pbd" style="background-color:#fffaf0;"></div> | |||
==See Also== | |||
*[[Actin 3D structures|Actin 3D structures]] | |||
*[[Profilin|Profilin]] | |||
*[[Profilin 3D Structures|Profilin 3D Structures]] | |||
*[[Vasodilator-stimulated phosphoprotein|Vasodilator-stimulated phosphoprotein]] | |||
== References == | |||
== | <references/> | ||
__TOC__ | |||
</StructureSection> | |||
== | |||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: Large Structures]] | |||
[[Category: Oryctolagus cuniculus]] | [[Category: Oryctolagus cuniculus]] | ||
[[Category: Dominguez R]] | |||
[[Category: Dominguez | [[Category: Ferron F]] | ||
[[Category: Ferron | [[Category: Rebowski G]] | ||
[[Category: Rebowski | |||
Latest revision as of 13:58, 30 August 2023
Ternary complex of profilin-actin with the poly-PRO-GAB domain of VASP*Ternary complex of profilin-actin with the poly-PRO-GAB domain of VASP*
Structural highlights
DiseasePROF1_HUMAN Defects in PFN1 are the cause of amyotrophic lateral sclerosis 18 (ALS18) [MIM:614808. A neurodegenerative disorder affecting upper motor neurons in the brain and lower motor neurons in the brain stem and spinal cord, resulting in fatal paralysis. Sensory abnormalities are absent. The pathologic hallmarks of the disease include pallor of the corticospinal tract due to loss of motor neurons, presence of ubiquitin-positive inclusions within surviving motor neurons, and deposition of pathologic aggregates. The etiology of amyotrophic lateral sclerosis is likely to be multifactorial, involving both genetic and environmental factors. The disease is inherited in 5-10% of the cases.[1] FunctionPROF1_HUMAN Binds to actin and affects the structure of the cytoskeleton. At high concentrations, profilin prevents the polymerization of actin, whereas it enhances it at low concentrations. By binding to PIP2, it inhibits the formation of IP3 and DG. Inhibits androgen receptor (AR) and HTT aggregation and binding of G-actin is essential for its inhibition of AR.[2] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedCells sustain high rates of actin filament elongation by maintaining a large pool of actin monomers above the critical concentration for polymerization. Profilin-actin complexes constitute the largest fraction of polymerization-competent actin monomers. Filament elongation factors such as Ena/VASP and formin catalyze the transition of profilin-actin from the cellular pool onto the barbed end of growing filaments. The molecular bases of this process are poorly understood. Here we present structural and energetic evidence for two consecutive steps of the elongation mechanism: the recruitment of profilin-actin by the last poly-Pro segment of vasodilator-stimulated phosphoprotein (VASP) and the binding of profilin-actin simultaneously to this poly-Pro and to the G-actin-binding (GAB) domain of VASP. The actin monomer bound at the GAB domain is proposed to be in position to join the barbed end of the growing filament concurrently with the release of profilin. Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP.,Ferron F, Rebowski G, Lee SH, Dominguez R EMBO J. 2007 Oct 31;26(21):4597-606. Epub 2007 Oct 4. PMID:17914456[3] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||