Glucocorticoid receptor
Function
Glucocorticoid receptor (GCR) is the nuclear receptor binding cortisol and glucocorticoids. GCR regulates genes involved in development, metabolism and immune response. GCR interacts with nuclear receptor coactivator 2 (NCOA2) which promotes DNA transcription by acylating histones[1].
Relevance
The glucocorticoid-bound GCR can either up-regulate the expression of anti-inflammatory proteins (transactivation) or repress the expression of pro-inflammatory proteins (transrepression)[2].
Structural highlights
GCR contains 5 domains: N terminal regulatory domain; DNA-binding domain (DBD) residues 417-506; hinge region; ligand-binding domain (LBD) residues 521-777 and C terminal domain.
3D structures of glucocorticoid receptor
Glucocorticoid receptor 3D structures
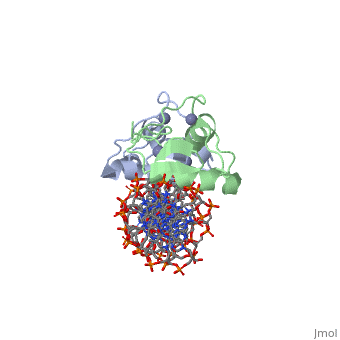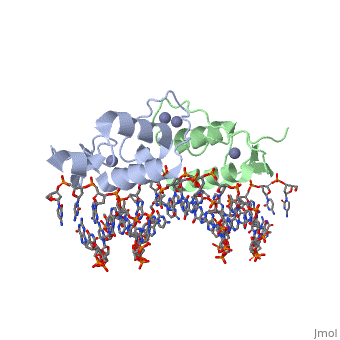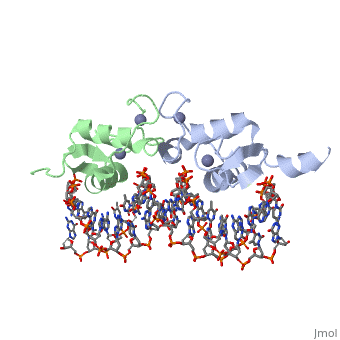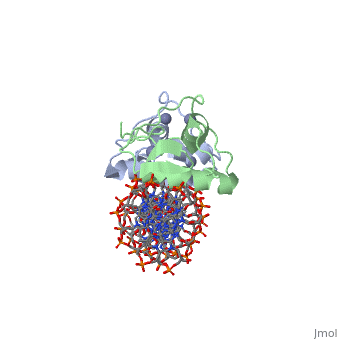
Rat glucocorticoid receptor DNA-binding domain dimer complex with DNA and Zn+2 ions (grey) (PDB entry 3g6r)
Drag the structure with the mouse to rotate 3D structures of glucocorticoid receptor
3D structures of glucocorticoid receptor Updated on 10-July-2019
References
References - ↑ Lu NZ, Wardell SE, Burnstein KL, Defranco D, Fuller PJ, Giguere V, Hochberg RB, McKay L, Renoir JM, Weigel NL, Wilson EM, McDonnell DP, Cidlowski JA. International Union of Pharmacology. LXV. The pharmacology and classification of the nuclear receptor superfamily: glucocorticoid, mineralocorticoid, progesterone, and androgen receptors. Pharmacol Rev. 2006 Dec;58(4):782-97. PMID:17132855 doi:http://dx.doi.org/10.1124/pr.58.4.9
- ↑ Finsterwald C, Alberini CM. Stress and glucocorticoid receptor-dependent mechanisms in long-term memory: from adaptive responses to psychopathologies. Neurobiol Learn Mem. 2014 Jul;112:17-29. doi: 10.1016/j.nlm.2013.09.017. Epub, 2013 Oct 7. PMID:24113652 doi:http://dx.doi.org/10.1016/j.nlm.2013.09.017
