Tom Sandbox

| |||||||
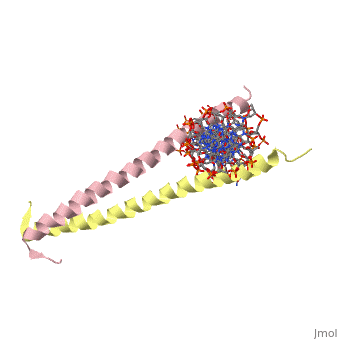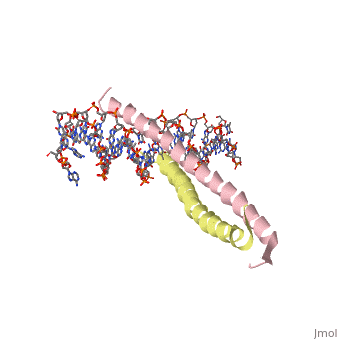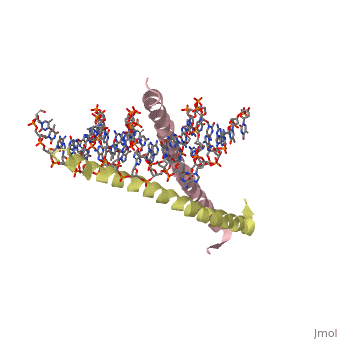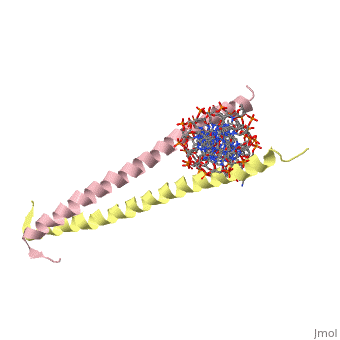
| 2zta, resolution 1.80Å () | |||||||
|---|---|---|---|---|---|---|---|
| Non-Standard Residues: | |||||||
| |||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
GCN4 - The Leucine ZipperGCN4 - The Leucine Zipper
Blah blah information about leucine zipper GCN4.
| |||||||||
| 1ysa, resolution 2.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
StructureStructure
GCN4 (PDB 2zta by itself, 1ysa bound to DNA) is a eukaryotic transcription factor first isolated from Saccharomyces cerevisiae, also known as Baker's Yeast. CITE CITE CITE It is composed of two identical 52 residue alpha helix chains that grouped together to form a dimer. The dimer binds through interlocking leucine amino acids in the C terminal ends, while pinching in on the major groove of DNA in the N terminal end. The X-ray structure of the 33-residue polypeptide corresponding to the leucine zipper of GCN4 was determined by Peter Kim and Thomas Alber[1].
BindingBinding
The Leucine ZipperThe Leucine Zipper
Binding with DNABinding with DNA
FunctionFunction
See AlsoSee Also
ReferenceReference
- ↑ Voet, Donald; Voet, Judith G.; Pratt, Charlotte W. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd Ed. Hoboken, NJ: Wiley, 2008.