Tom Sandbox

| |||||||
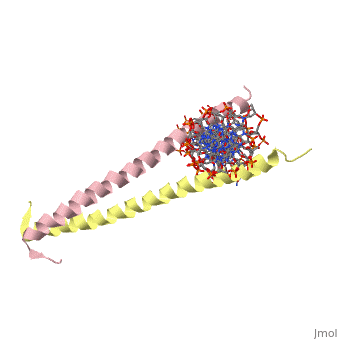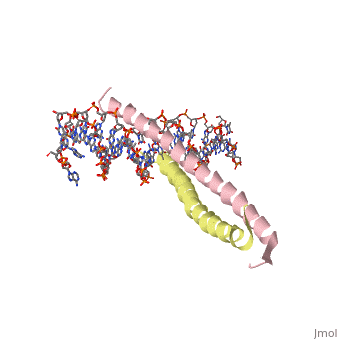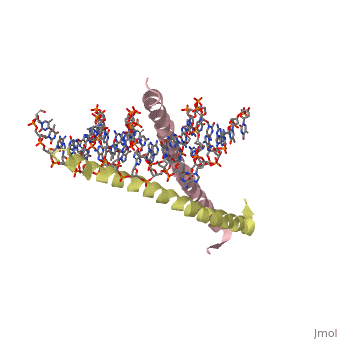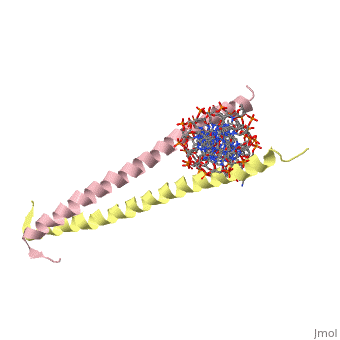
| 2zta, resolution 1.80Å () | |||||||
|---|---|---|---|---|---|---|---|
| Non-Standard Residues: | |||||||
| |||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
GCN4 - The Leucine ZipperGCN4 - The Leucine Zipper
GCN4 (PDB 2zta by itself, 1ysa bound to DNA) is a eukaryotic transcription factor first isolated from Saccharomyces cerevisiae, also known as Baker's Yeast. It is required for the general control of amino acid levels in yeast and binds upstream of several amino acid bio-synthetic genes.[1] Its primary function is to act as one of the main response factors for amino acid starvation by initiating transcription, however the exact mechanism of initiation is still being debated (see Binding with DNA).
GCN4 is known as a bZip protein after its basic region and leucine zipper domains. The first 'leucine zipper' model was coined by Landshulz et al. in 1988 after their initial X-Ray crystallography structure was solved. Today, while the name has stayed the same, we no longer view the leucine binding region in an inter-collated manner like the teeth of a zipper, but as Leucines meeting face to face [2]. (see heptad repeat section) the Leucines are represented as red and the Valines are shown as orange. A close up of the leucine-leucine pairing can be seen . GCN4 binds to promoter regions AP-1 and ATF/CREB to induce transcription via the C terminal basic residues of the two symmetric alpha helices.[3]
Hydrophobic StabilityHydrophobic Stability
GCN4 is composed of two identical 58 residue alpha helix chains that grouped together to form a parallel coiled-coil dimer. Roughly 33 of the 58 total residues are located in the Leucine Zipper region. The dimer binds through interlocking leucine amino acids and near the C terminus, while pinching in on the major groove of DNA in the N terminal end via basic residues. The aforementioned image shows how the hydrophobic regions are between the helices, while the hydrophobic regions protrude outwards.
| |||||||||
| 1ysa, resolution 2.90Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Zipper and Binding DomainsZipper and Binding Domains
The two main domains are labeled the . [4] Here the acidic region is represented in red and the basic region in blue. The basic residues are the reason the class of binding interactions is commonly referred to as bZIP or basic region leucine zipper proteins[5]. The basic region at the N-terminal of the two chains clamps in on the DNA in a scissor-like manner, making contact with both the and the of DNA.
Binding with DNABinding with DNA
The basic region binding domain of GCN4 inserts itself into the of the DNA. This causes a bend in both the AP-1 and ATF/CREB binding sites of DNA[6]. AP-1 refers to a group of activator proteins that bind to the same 9 residue semi-palindromic region for transcription activation. ATF/CREB refers to a fully palindromic region. The AP-1 site sequence is 5′-ATGACTCAT-3′, and the ATF/CREB site is 5′-ATGACGTCAT-3′[7]. The bending mechanism was determined by adding fluorophores to the ends of U shaped manufactured DNA segments with the specific binding sites AP-1 or ATF/CREB inserted into the center. The shift in the fluorophore positioning showed the effect of bending on the DNA due to GCN4 binding. In total, in the complex with GCN4-bZIP, the ATF/CREB site is bent by (25 ± 2)° and the AP-1 site by (20 ± 2)° toward the minor groove.[6] This is thought to be the mechanism for inducing transcription, however the actual bending of DNA is still debated.[8][9]
Heptad RepeatHeptad Repeat
GCN4 is said to have a heptad repeat, or seven unit repeated sequence. Here the heptad repeat is a leucine, every seven units. When the structure of the alpha helices is taken into account, it is clear that each leucine is being stacked on top of the last one, every seven units. This creates the zipper-like repeated projections from the two polypeptide chains. Below we see a pictorial representation of the sequence of amino acids as we look down the length of the coiled coil structure.

Image 1:A pictorial representation of the heptad repeat between the two subunits in a coiled-coil conformation. By following through the wheels alphabetically, it is clear how the leucines will stack every 7 units. The apostrophe denotes the diference between the two polypeptide subunits.(from Kgutwin in Wikipedia Commons http://commons.wikimedia.org/wiki/File:Coiledcoil-wheelcartoon.gif)
At every d and d' we have a Leucine, while at the a and a' locations we typically see valine. [5] In the opposite positions one tends to see more charged and polar residues. The Leucine and Valine repeats create a strongly hydrophobic region between the two alpha helices. This clearly shows the relationship between coils and shown below, we can see that for the two helices to be symmetrical, the Leucines must not in fact be oriented as a zipper, but on the same levels.

Image 2: The figure above comes from the paper - Secondary Structure of a Leucine Zipper Determined by NMR.[2] Figure A represents the interdigitated zipper model and figure B shows the coiled coil symmetric model that is now the accepted structure.
The Leucine Zipper Nano-TweezerThe Leucine Zipper Nano-Tweezer
The X-ray structure of the 33-residue polypeptide corresponding to the leucine zipper of GCN4 was determined by Peter Kim and Thomas Alber in 1991[10]. As stated above, the leucines themselves come on every other level of the alpha helix and do not actually interchange one over the other like a zipper, but instead make side to side contact. This was noted in 1990 due to the symmetric nature of the two subunits. If the leucines showed interdigitation the subunits would be asymmetric [2].
The Leucine Zipper of GCN4, as expected of a protein, operates under a specific pH. It has been shown that at lower pH values the zipper will reversibly protenate and open up. This occurs because it loses its hydrophobic stability, protonating its polypeptide chains. Researchers are looking into this opening and closing reaction to be used purposefully as a form of nano-tweezers to grab onto and hold very-very small particles. [4] A diagram of this mechanism is shown below.

Image 3: A schematic representation of the Leucine Zipper acting as nano-tweezers under different pH conditions[4].
See AlsoSee Also
ReferenceReference
- ↑ Arndt K, Fink GR. GCN4 protein, a positive transcription factor in yeast, binds general control promoters at all 5' TGACTC 3' sequences. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8516-20. PMID:3464968
- ↑ 2.0 2.1 2.2 Oas, T. G.; McIntosh, L. P.; O'Shea, E. K.; Dahlquist, F. W.; and Kim, P. S. Biochemistry 1990 29 (12), 2891-2894
- ↑ Hope, I. A.; Struhl,K. Cell, Volume 46, Issue 6, 12 September 1986, Pages 885-894
- ↑ 4.0 4.1 4.2 Sharma, G.; Rege, K.; Budil, D. E.; Yarmush, M. L.; Mavroidis, C. Int J Nanomedicine. 2008 December; 3(4): 505–521.
- ↑ 5.0 5.1 Voet, Donald; Voet, Judith G.; Pratt, Charlotte W. Fundamentals of Biochemistry: Life at the Molecular Level. 3rd Ed. Hoboken, NJ: Wiley, 2008.
- ↑ 6.0 6.1 Dragan AI, Liu Y, Makeyeva EN, Privalov PL. DNA-binding domain of GCN4 induces bending of both the ATF/CREB and AP-1 binding sites of DNA. Nucleic Acids Res. 2004 Sep 30;32(17):5192-7. Print 2004. PMID:15459288 doi:10.1093/nar/gkh854
- ↑ Hockings, S. C.; Kahn, J. D.; Crothers, D. M. PNAS February 17, 1998 vol. 95 no. 4 1410-1415
- ↑ Strauss-Soukup JK, Maher LJ 3rd. DNA bending by GCN4 mutants bearing cationic residues. Biochemistry. 1997 Aug 19;36(33):10026-32. PMID:9254597 doi:10.1021/bi970215u
- ↑ Benevides JM, Li T, Lu XJ, Srinivasan AR, Olson WK, Weiss MA, Thomas GJ Jr. Protein-directed DNA structure II. Raman spectroscopy of a leucine zipper bZIP complex. Biochemistry. 2000 Jan 25;39(3):548-56. doi: 10.1021/bi990053x. PMID:10642179 doi:http://dx.doi.org/10.1021/bi990053x
- ↑ O'Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991 Oct 25;254(5031):539-44. PMID:1948029
- ↑ Daniel Zaidman, Paul Gehrtz, Mihajlo Filep, Daren Fearon, Ronen Gabizon, Alice Douangamath, Jaime Prilusky, Shirly Duberstein, Galit Cohen, C. David Owen, Efrat Resnick, Claire Strain-Damerell, Petra Lukacik, Haim Barr, Martin A. Walsh, Frank von Delft, Nir London, An automatic pipeline for the design of irreversible derivatives identifies a potent SARS-CoV-2 Mpro inhibitor, Cell Chemical Biology, 2021 doi:https://dx.doi.org/10.1016/j.chembiol.2021.05.018