Succinate Dehydrogenase
Succinate DehydrogenaseSuccinate Dehydrogenase
Succinate dehydrogenase (PDB = 2wdv with empty ubiquinone binding site; PBD = 1nek with ubiquinone bound), also called succinate-coenzyme Q reductase (SQR) or Complex II, is a tetrameric enzyme found in the cell membrane of some bacteria and the inner mitochondrial membrane of mammalian cells. It is classified as an α+β protein, as it contains segregated regions of α helices and antiparallel β sheets. It is involved in two aspects of digestion; it catalyzes the oxidation of succinate to fumarate in the citric acid cycle by simultaneously reducing ubiquinone to ubiquinol in the electron transport chain [1].
| |||||||||
| 2wdv, resolution 3.20Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , , , , , | ||||||||
| Activity: | Succinate dehydrogenase (ubiquinone), with EC number 1.3.5.1 | ||||||||
| Related: | 2wdq, 2wdr, 1nek, 1nen, 2acz | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Structure:Structure:
The tetramer is composed of two hydrophilic and two hydrophobic subunits. The hydrophilic subunits are named SdhA and SdhB; the former is a flavoprotein containing a covalently-bound FAD cofactor a binding site for succinate, while the latter is Fe-S protein bearing the three iron-sulfur clusters 2Fe-2S, 3Fe-4S, and 4Fe-4S. The hydrophobic subunits, termed SdhC and SdhD, anchor the protein in the mitochondrial membrane and formally comprise cytochrome b [2] and [3]. This cytochrome contains six transmembrane α-helices, a heme b group, and a binding site for ubiquinone located in a space bounded by SdhB, SdhC, and SdhD [4] and [5].
| |||||||||
| 1nek, resolution 2.60Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , , , , , , , , | ||||||||
| Gene: | SDHA OR B0723 OR Z0877 OR ECS0748 (Escherichia coli), SDHB OR B0724 (Escherichia coli), SDHC OR CYBA OR B0721 OR Z0875 OR ECS0746 (Escherichia coli), SDHD OR B0722 (Escherichia coli) | ||||||||
| Related: | 1nen | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Binding sites:Binding sites:
SuccinateSuccinate
The binding site for succinate, in which the stereospecific dehydrogenation of succinate to fumarate is catalyzed, is located entirely on SdhA. Residues Thr254, His354, and Arg399 stabilize the substrate with hydrogen bonding, while FAD removes the electrons and carries them to the first iron-sulfur cluster, 2Fe-2S, of SdhB [6]. During this transfer, FAD is reduced to FADH2 [7]
UbiquinoneUbiquinone
The binding site for ubiquinone, in which the substrate is reduced to ubiquinol, is bordered by subunits B, C, and D. Residues His207 of SdhB, Ser27 and Arg31 of SdhC, and Tyr83 of SdhD stabilize ubiquinone, while residues Pro160, Trp163, Trp164, and Ile209 of SdhB and Ser27 and Ile28 of SdhC provide the necessary hydrophobic environment that stabilizes the ring [8].
Mechanisms:Mechanisms:
Succinate oxidationSuccinate oxidation
The exact mechanism for the oxidation of succinate to fumarate has not yet been elucidated. The initial deprotonation may be performed by FAD, Glu255, Arg286, or His242 of SdhA, and the following elimination may be a concerted E2 or E1cb elimination. In the concerted mechanism, the α-carbon is deprotonated by a base as FAD removes a hydride from the β-carbon; this is shown in image 1 [9].
Image 1: Oxidation of succinate to fumarate through E2 elimination
In the proposed E1cb mechanism, the deprotonation leads to the formation of an enolate intermediate; FAD then removes the hydride, as shown in Image 2 [10].
Image 2: Oxidation of succinate to fumarate via E1cb elimination
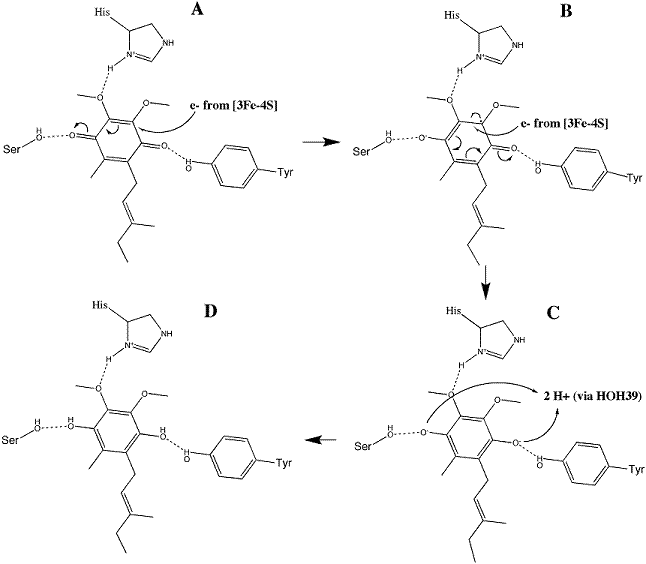
Ubiquinone reductionUbiquinone reduction
Ubiquinone is initially oriented in the active site such that the O1 carbonyl group interacts with Tyr83 of SdhD via hydrogen bonding. The electrons removed during the oxidation reaction are conveyed through the iron-sulfur clusters to 3Fe-4S; their presence on that cluster stimulates the substrate to reorient so that a second hydrogen bond between the O4 carbonyl group and Ser27 of SdhC may form. The electrons are transferred to the substrate individually, with the addition of the first producing a radical semiquinone and the second completing the reduction to ubiquinol. This mechanism is illustrated in image 3 [11].
Image 3: Reduction of ubiquinone to ubiquinol
References:References:
- ↑ Oyedotun KS, Lemire BD. The quaternary structure of the Saccharomyces cerevisiae succinate dehydrogenase. Homology modeling, cofactor docking, and molecular dynamics simulation studies. J Biol Chem. 2004 Mar 5;279(10):9424-31. Epub 2003 Dec 12. PMID:14672929 doi:10.1074/jbc.M311876200
- ↑ Tomitsuka E, Hirawake H, Goto Y, Taniwaki M, Harada S, Kita K. Direct evidence for two distinct forms of the flavoprotein subunit of human mitochondrial complex II (succinate-ubiquinone reductase). J Biochem. 2003 Aug;134(2):191-5. PMID:12966066
- ↑ Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003 Jan 31;299(5607):700-4. PMID:12560550 doi:10.1126/science.1079605
- ↑ Yankovskaya V, Horsefield R, Tornroth S, Luna-Chavez C, Miyoshi H, Leger C, Byrne B, Cecchini G, Iwata S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003 Jan 31;299(5607):700-4. PMID:12560550 doi:10.1126/science.1079605
- ↑ Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Omura S, Byrne B, Cecchini G, Iwata S. Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction. J Biol Chem. 2006 Mar 17;281(11):7309-16. Epub 2005 Dec 27. PMID:16407191 doi:http://dx.doi.org/10.1074/jbc.M508173200
- ↑ Kenney WC. The reaction of N-ethylmaleimide at the active site of succinate dehydrogenase. J Biol Chem. 1975 Apr 25;250(8):3089-94. PMID:235539
- ↑ ISBN:978-0-470-12930-2
- ↑ Horsefield R, Yankovskaya V, Sexton G, Whittingham W, Shiomi K, Omura S, Byrne B, Cecchini G, Iwata S. Structural and computational analysis of the quinone-binding site of complex II (succinate-ubiquinone oxidoreductase): a mechanism of electron transfer and proton conduction during ubiquinone reduction. J Biol Chem. 2006 Mar 17;281(11):7309-16. Epub 2005 Dec 27. PMID:16407191 doi:http://dx.doi.org/10.1074/jbc.M508173200
- ↑ Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b. J Biol Chem. 2006 Oct 27;281(43):32310-7. Epub 2006 Sep 1. PMID:16950775 doi:10.1074/jbc.M607476200
- ↑ Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b. J Biol Chem. 2006 Oct 27;281(43):32310-7. Epub 2006 Sep 1. PMID:16950775 doi:10.1074/jbc.M607476200
- ↑ Tran QM, Rothery RA, Maklashina E, Cecchini G, Weiner JH. The quinone binding site in Escherichia coli succinate dehydrogenase is required for electron transfer to the heme b. J Biol Chem. 2006 Oct 27;281(43):32310-7. Epub 2006 Sep 1. PMID:16950775 doi:10.1074/jbc.M607476200