AlphaFold2 examples from CASP 14
Prediction of protein structures from amino acid sequences, theoretical modeling, has been extremely challenging. In 2020, breakthrough success was achieved by AlphaFold2[1], a project of DeepMind. For an overview of this breakthrough, documented by the bi-annual prediction competition CASP, please see 2020: CASP 14. Below are illustrated two examples of predictions from that competition.
Free Modeling ResultsMore than 100 domains were provided as prediction targets in CASP 14. 14 of these were in the most difficult category, free modeling ("FM"), meaning that no informative homology modeling templates existed. For 8 of these (57%), AlphaFold2's predictions achieved GDT_TS scores of 87-93 (median 88.5). For those 8, GDT_TS of the second best predictions were 43-76 (median 66). Two cases will be analyzed below. First, SARS-CoV-2 ORF8[2], a 92-residue FM domain where AlphaFold2's GDT_TS was 87, and the second best was 43 (by the group of Xian Ming Pan)[3], the largest difference between 1st and 2nd predictions among the FM targets. It is further unusual because two independently-determined X-ray crystallographic structures were subsequently published. Inspiration for this case came from the discussion by Rubiera[4]. Second, the longest domain in the FM category, 404 residues. This domain is part of the 2,180-residue RNA polymerase of a bacteriophage, some of whose group members are prevalent in the human gut[5]. Eight of the CASP 14 FM target domains are parts of this protein, 6vr4. For the 404-residue domain T1037, AlphaFold2 achieved GDT_TS of 88, and the second best prediction, 63 (by Seok-refine)[6]. Among the 14 FM targets, the second-longest has 276 residues, the median 132, and the shortest, 92. SARS-CoV-2 ORF8Our first example is SARS-CoV-2 protein ORF8, a protein that contributes to virulence in COVID-19[2]. CASP 14 classified ORF8 as a "free modeling" (FM) target[7], meaning that there were no adequate empirical templates for homology modeling. This was easily confirmed. When the amino acid sequence of ORF8 is submitted to Swiss Model, it reports the best templates for homology modeling. When the two empirical models that were not available during CASP 14 are excluded (7jtl and 7jx6), the best template offered, chain B of 3afc, covers only 36% of the length of ORF8 at 13.2% sequence identity, with a 4-residue untemplated gap in the sequence alignment. This template would not be adequate for constructing a useful model. X-Ray Structures for ORF8The quality of predictions for the structure of ORF8 are judged by comparison with X-ray crystallographic empirical models which were not available to the groups making predictions. Shortly after the CASP 14 competition (summer 2020), two X-ray crystal structures were reported for ORF8: 7jtl released August 26, 2020, and 7jx6, released September 23, 2020. The resolutions are 2.0 and 1.6 Å respectively, and both have worse than average Rfree values.
from the higher resolution X-ray structure, 7jx6. These chains form disulfide-linked dimers, and the dimers form higher order multimers[2] (not shown). Notice that the amino and carboxy ends of the chain come together to form two parallel beta strands of a beta sheet. Also notice that there are 3 disulfide bonds. An accurate prediction would include both of these features. [8]. The only substantial disagreement is for a large surface loop, sequence range 48-57. See the Table I below for RMSD values. ORF8 is not a novel foldLess than 2% of new empirically-determined structures have novel folds; that is, folds not aready represented in the PDB[9]. When chain A of 7jx6 was submitted to Dali[10] (February, 2021), the top hit was the N-terminal domain of the two domains in 5a2f, the CD166 human cell surface receptor involved in activation of T lymphocytes. The Z-score was 7.1, and 88 alpha carbons superposed with RMSD 3.2 Å. Swiss-PdbViewer obtained RMSD 1.95 Å for 48 alpha carbons[11]. Dali reported the identity as 6% in its structure-based sequence alignment. Sequence alignment by MAFFT[12] obtained 18% sequence identity using more and larger gaps. [13] is not as close as for AlphaFold2's prediction, but is closer than the 2nd best prediction (see Table I below). In conclusion, ORF8 does not have a novel fold[14]. AlphaFold2 Prediction for ORF8The quality of a prediction in CASP is judged, in large part, by the Global Distance Test Total Score, GDT_TS. AlphaFold2's predicted structure[15] has a GDT_TS score of 87. (A score of 0 is meaningless, and a score of 100 means perfect agreement with an X-ray crystal structure.) 87 means [8]. The structure predicted by AlphaFold2 is almost as close to the X-ray crystallographic model 7jx6 as is the independently-determined X-ray structure 7jtl. AlphaFold2 predicted the positions of 92 amino acids. (CASP 14 excluded residues 48-59, a 12-residue surface loop, from the target residues[7].) See Table I below for RMSD values. The prediction was largely accurate regarding salt bridges and cation-pi interactions (see Tables III and IV below).
Second Best Prediction for ORF8In CASP 14, 70 research groups and 42 automated servers predicted structures for ORF8. The median GDT_TS score for all 112 predictions was 26. AlphaFold2 made the best prediction (GDT_TS 87). [8], with GDT_TS 43 (see Table I above). The fold and topology were predicted correctly, but the details are far less accurate than those in AlphaFold2's prediction. The 2nd best prediction has no disulfide bonds. This prediction was largely incorrect regarding salt bridges and cation-pi interactions (see Tables III and IV below). Third Best Prediction for ORF8The third best prediction for ORF8 was by the Perez Lab, with GDT_TS 33 (see Table I above). It correctly predicted the parallel beta strands formed by the amino and carboxy terminal ends of the chain. [19]. This prediction has no disulfide bonds. The salt bridge Arg86:Asp98 is correctly predicted, along with two incorrectly predicted salt bridges. Top Prediction by an Automated ServerAmong predictions by automated servers for all ~100 CASP 14 targets, the top ranking server was QUARK from the Yang Zhang group (Univ. Michigan). For ORF8, the Zhang-TBM server made the best server prediction with a GDT_TS of 27. (The prediction by QUARK was almost as good, GDT_TS 26.) The prediction has the two chain termini not parallel, and the amino terminus is not a beta strand, differing in both respects from the X-ray model. Also, no disulfide bonds are predicted. The salt bridge Arg86:Asp98 is correctly predicted, along with several incorrectly predicted salt bridges. The structural superposition is very poor and is not shown. Baker Rosetta Server Prediction for ORF8Among predictions for all ~100 CASP 14 targets, the group of David Baker ranked second. The Rosetta Server of the Baker group ranked 18th overall, but was the 4th ranked server[20]. For ORF8, the Rosetta Server prediction GDT_TS was 26, a bit better than the median of 23. The Rosetta Server's prediction for ORF8 has the two termini far apart (Cα 13 Å or farther apart), a substantial difference from the X-ray structure (Cα mostly ~5 Å apart). It predicts two disulfide bonds, but neither matches the pairs of Cys residues in the actual disulfide bonds. The salt bridge Arg86:Asp98 is correctly predicted, along with one incorrectly predicted salt bridge. The structural superposition is very poor and is not shown. ORF8 Sidechain Prediction AccuracyJump below to #ORF8 Sidechain Accuracy Phage RNA polymerase T1037Our second example is a 404-residue domain within a 2,180 residue RNA polymerase, 6vr4, gp66 from a bacteriophage of the crAss-like group, some members of which are prevalent in the human gut[5]. One known host for the target-containing phage is gram-negative, aerobic bacteria Cellulphaga baltica isolated from marine microalgae[5][21]. This RNA polymerase is packaged in the virion, and delivered into the host cell, where it transcribes early phage genes[5]. CASP 14 classified this domain, coded T1037, as a free modeling ("FM") target[7], meaning that no informative templates existed in the PDB. X-ray crystal structureThe crystallographic structure, not available to the prediction teams during the CASP competition, is 6vr4, with resolution 3.5 Å, and an Rfree "reliability" of "much better than average for this resolution" (according to FirstGlance in Jmol). The termini of the chain are far apart from each other (~85 Å), and there are no disulfide bonds. The asymmetric unit contains 2 chains. The reference structure was taken from chain B because it has a lower average temperature factor than chain A. The two chains are nearly identical (see Table II below). The CASP 14 target T1037 sequence of 404 residues begins at sequence number 337 and ends at 901, a span of 565 residues. The target sequence is 404 residues because it excludes residues 370-530 (length 161), which form a different domain. . Here is the 404-residue target sequence with the intervening domain (excluded from the CASP target) . (length 404 residues) consists of residues 337-369 + 531-901 of 6vr4 (taken from chain B). It is an 45% helices, 19% beta strands , and 37% loops and turns. The N- and C-termini are 10 Å apart, and there are no cysteines (thus no disulfide bonds). T1037 contains several known fold fragmentsThe X-ray structure of T1037 (404 residues from 6vr4) was submitted to Dali[10] in March, 2021. Among the ~1,000 hits with Z ≥ 2.0, there were 152 with lengths ≥ 400 residues, and 224 with lengths ≥ 300, long enough that a superposition with the majority of T1037 would not be precluded. Among all hits, the largest number of aligned residues was 140/404 (35%) with RMSD 11.7 Å. The second largest was 127/404 (31%), RMSD 7.7 Å. Thus, no single structure in the PDB superposed with more than 35% of T1037. However, several of the Dali hits superposed with non-overlapping core fragments of 6vr4[22]:
Liisa Holm[10][23] stated: "T1037 has a homologous template in the PDB. The parent structure of T1037, phage RNA polymerase (6vr4, 2166 amino acids), is homologous to the RNAi polymerase from Neurospora crassa (2j7nA, 934 amino acids)[5]. Dali aligns them over 564 residues with an RMSD of 4.8 A. 115 residues of the common core are in the T1037 substructure. Several long insertions in T1037/6vr4 relative to 2j7n (chain A) form subdomains, which point outwards from the common core. Similar massive adaptation of the common core is seen, for example, in the glucosyltransferase 1 family[24]." The FATCAT Server reported that in order to superpose 150 residues (37% of 404) of T1037 with the closest structure in the PDB, 3 twists at hinges were required, after which an RMSD of 3.1 Å was achieved. For a 200-residue superposition (50% of 404), the best results after 3 twists had an RMSD of 5.4 Å. AlphaFold2 prediction for T1037[8]. 91% of the 404 alpha carbons can be aligned with RMSD 1.0 Å. (GDT_TS 88; see Table II below for details).
Second Best Prediction for T1037Despite its impressive GDT_TS of 63, [8] than the prediction of AlphaFold2. (The second best prediction was by Seok-refine, from the group of Chaok Seok, Seoul National University.) Calculating GDT_TSPlease see #GDT_TS Calculations.
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
ORF8 Sidechain AccuracyORF8 Sidechain Accuracy
AlphaFold2's predictions for sidechain positions seem fairly good, while sidechain positions in the 2nd best prediction seem poor. This conclusion is based on three types of observations:
- Table I gives RMSD values for all atoms, which is one indication of sidechain accuracy.
- Prediction of salt bridges and cation-pi interactions.
- Visualization of the distributions of charges on the surfaces.
Salt Bridges and Cation-Pi InteractionsSalt Bridges and Cation-Pi Interactions
- AlphaFold2's prediction was correct for 4/5 interactions, with one incorrect interaction.
- AlphaFold2's prediction was correct for one of two salt bridges, and predicted no incorrect salt bridges.
- AlphaFold2's prediction was correct for three of three cation-pi interactions, but predicted one incorrect interaction.
- The 2nd best prediction was correct for 1/5 interactions, with 2 incorrect interactions.
- The 2nd best prediction was correct for one of two salt bridges, but predicted two incorrect salt bridges.
- The 2nd best prediction failed to predict any of the three cation-pi interactions, predicting zero interactions.
| 7JX6 | 7JTL | AlphaFold2 | 2nd Best |
|---|---|---|---|
| R101:D112 (AB) | R101:D113 (AB) | R86:D98 | R86:D98 |
| R115:D119 (AB) | R115:D119 (AB) | – | R100:E4 |
| K44:E59 (AB) | K44:E59 (AB) | K29:E44 | – |
| – | – | – | K78:E77 |
- Bridges in the same row are identical (except for red residues). Subtract 15 from the sequence numbers in the X-ray structures for the equivalent sequence numbers in the predictions.
- Black: Shortest sidechain nitrogen to sidechain oxygen distance ≤4.0 Å.
- Gray: Shortest sidechain nitrogen to sidechain oxygen distance 4.4 to 4.8 Å.
- –: Shortest sidechain nitrogen to sidechain oxygen distance 6 to 16 Å.
- (AB): The two chains in each X-ray model.
- Italics: erroneous prediction.
| 7JX6 | 7JTL | AlphaFold2 | 2nd Best |
|---|---|---|---|
| R101:Y46+Y108 (AB) | R101:Y46+Y108 (AB) | R86:Y31+Y96 | – |
| K44:F108 (B) | K44:F108 (AB) | K29:F93 | – |
| – | – | K79:F105 | – |
- All interactions listed are deemed energetically significant by the CaPTURE Server.
- Interactions in the same row are identical. Subtract 15 from the sequence numbers in the X-ray structures for the equivalent sequence numbers in the predictions.
- Italics: erroneous prediction.
- The 2nd best prediction has no cation-pi interactions.
- (AB): The two chains in each X-ray model.
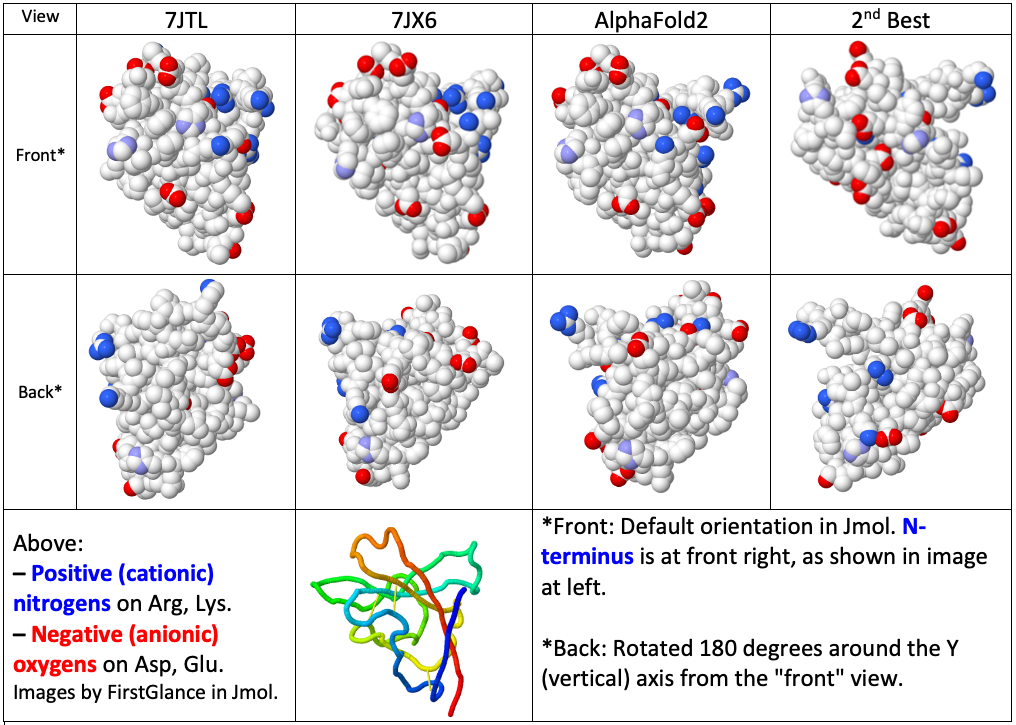
Visualization of Surface Charge DistributionsVisualization of Surface Charge Distributions
The distributions of surface charges are in good agreement between AlphaFold2's prediction and the two crystal structures, which agree with each other. The distribution in the 2nd best prediction has several discrepancies with the other three models.
GDT_TS CalculationsGDT_TS Calculations
GDT_TS values for predictions are taken from CASP 14 results. The reference structure for the CASP 14 GDT_TS values was 92 alpha carbons of 7JTL[7], since the CASP 14 target had only 92 residues[7].
GDT_TS values for 7JTL and 5A2F vs. 7JX6 chain A were calculated using the AS2TS server of Adam Zemla[25]. See instructions for Calculating GDT_TS. GDT_TS values were corrected for 92 residues (not 104) because the CASP 14 target had only 92 residues[7].
For comparison, CASP 14 reported GDT_TS 86.96 for the AlphaFold2 prediction, while the AS2TS server calculated GDT_TS 85.87 vs. 7jx6 chain A, and 88.32 vs. 7JTL chain A. (These results were corrected for 90/92 and 91/92 residues, respectively.) Thus, there appears to be some unidentified minor discrepancy between the GDT_TS calculations of CASP-14 vs. the method detailed at Calculating GDT_TS.
References & NotesReferences & Notes
- ↑ Senior AW, Evans R, Jumper J, Kirkpatrick J, Sifre L, Green T, Qin C, Zidek A, Nelson AWR, Bridgland A, Penedones H, Petersen S, Simonyan K, Crossan S, Kohli P, Jones DT, Silver D, Kavukcuoglu K, Hassabis D. Improved protein structure prediction using potentials from deep learning. Nature. 2020 Jan;577(7792):706-710. doi: 10.1038/s41586-019-1923-7. Epub 2020 Jan, 15. PMID:31942072 doi:http://dx.doi.org/10.1038/s41586-019-1923-7
- ↑ 2.0 2.1 2.2 Flower TG, Buffalo CZ, Hooy RM, Allaire M, Ren X, Hurley JH. Structure of SARS-CoV-2 ORF8, a rapidly evolving immune evasion protein. Proc Natl Acad Sci U S A. 2021 Jan 12;118(2). pii: 2021785118. doi:, 10.1073/pnas.2021785118. PMID:33361333 doi:http://dx.doi.org/10.1073/pnas.2021785118
- ↑ For SARS-CoV-2 ORF8, at the CASP 14 Table Browser, check T1064-D1 and press Show Results.
- ↑ CASP14: what Google DeepMind’s AlphaFold 2 really achieved, and what it means for protein folding, biology and bioinformatics, a blog post by Carlos Outeir al Rubiera, December 3, 2020.
- ↑ 5.0 5.1 5.2 5.3 5.4 Drobysheva AV, Panafidina SA, Kolesnik MV, Klimuk EI, Minakhin L, Yakunina MV, Borukhov S, Nilsson E, Holmfeldt K, Yutin N, Makarova KS, Koonin EV, Severinov KV, Leiman PG, Sokolova ML. Structure and function of virion RNA polymerase of a crAss-like phage. Nature. 2020 Nov 18. pii: 10.1038/s41586-020-2921-5. doi:, 10.1038/s41586-020-2921-5. PMID:33208949 doi:http://dx.doi.org/10.1038/s41586-020-2921-5
- ↑ For the phage RNA polymerase target, at the CASP 14 Table Browser, check T1037-D1 and press Show Results.
- ↑ 7.0 7.1 7.2 7.3 7.4 7.5 Summary, Definitions and Classifications of Domains for CASP 14.
- ↑ 8.0 8.1 8.2 8.3 8.4 8.5 8.6 Superposition by Swiss-PdbViewer's iterative magic fit. This starts with a sequence alignment-guided structural superposition, and then superposes subsets of the structures to minimize the RMSD. Eight intermediate structures were generated by the Theis Morph Server by linear interpolation.
- ↑ Cuff AL, Sillitoe I, Lewis T, Clegg AB, Rentzsch R, Furnham N, Pellegrini-Calace M, Jones D, Thornton J, Orengo CA. Extending CATH: increasing coverage of the protein structure universe and linking structure with function. Nucleic Acids Res. 2011 Jan;39(Database issue):D420-6. doi: 10.1093/nar/gkq1001. , Epub 2010 Nov 19. PMID:21097779 doi:http://dx.doi.org/10.1093/nar/gkq1001
- ↑ 10.0 10.1 10.2 Holm L. DALI and the persistence of protein shape. Protein Sci. 2020 Jan;29(1):128-140. doi: 10.1002/pro.3749. Epub 2019 Nov 5. PMID:31606894 doi:http://dx.doi.org/10.1002/pro.3749
- ↑ Using Swiss-PdbViewer's Fit from Selection with 102 residues selected from each structure, followed by Improve Fit.
- ↑ Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013 Apr;30(4):772-80. doi: 10.1093/molbev/mst010. Epub 2013 Jan, 16. PMID:23329690 doi:http://dx.doi.org/10.1093/molbev/mst010
- ↑ Structural superposition by Dali. Interpolation by the Yale Morph2 Server. Homogenization method: homology modeling. No minimization. This produced a 9-model file where model 1 was 7jx6, and models 2-9 were interpolations. 5a2f residues 28-133 were added as model 10 (black in the molecular scene).
- ↑ The interpretation of Dali's result to mean that ORF8 does not have a novel fold was kindly confirmed by Liisa Holm, personal communication to Eric Martz, February, 2021.
- ↑ Download AlphaFold2's predicted structure for ORF8 from T1064TS427_1-D1.pdb.
- ↑ 16.0 16.1 16.2 See #GDT_TS Calculations.
- ↑ See #ORF8 is not a novel fold.
- ↑ 18.0 18.1 Superposition by Swiss-PdbViewer's magic fit. This is a sequence alignment-guided structural superposition. Eight intermediate structures were generated by the Theis Morph Server by linear interpolation.
- ↑ Superposition by Swiss-PdbViewer's Explore Fragment Alternate Fits, which does not use sequence information. Eight intermediate structures were generated by the Theis Morph Server by linear interpolation.
- ↑ For all targets in CASP 14, the top two servers were QUARK and Zhang-server (which were not significantly different at a Z-score sum of 62.9), followed by Zhang-CEthreader (55.9) and BAKER-ROSETTASERVER (55.3).
- ↑ Johansen JE, Nielsen P, Sjoholm C. Description of Cellulophaga baltica gen. nov., sp. nov. and Cellulophaga fucicola gen. nov., sp. nov. and reclassification of [Cytophaga] lytica to Cellulophaga lytica gen. nov., comb. nov. Int J Syst Bacteriol. 1999 Jul;49 Pt 3:1231-40. doi: 10.1099/00207713-49-3-1231. PMID:10425785 doi:http://dx.doi.org/10.1099/00207713-49-3-1231
- ↑ These non-overlapping core fragments were kindly pointed out by Liisa Holm, March, 2021.
- ↑ Quoted with permission from Liisa Holm, March, 2021.
- ↑ Holm L, Sander C. Evolutionary link between glycogen phosphorylase and a DNA modifying enzyme. EMBO J. 1995 Apr 3;14(7):1287-93. PMID:7729407
- ↑ Zemla A. LGA: A method for finding 3D similarities in protein structures. Nucleic Acids Res. 2003 Jul 1;31(13):3370-4. doi: 10.1093/nar/gkg571. PMID:12824330 doi:http://dx.doi.org/10.1093/nar/gkg571