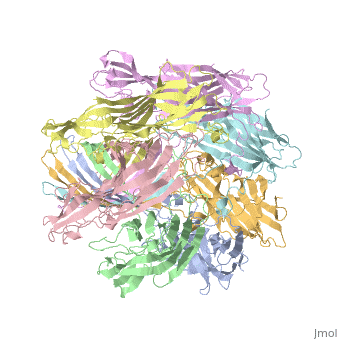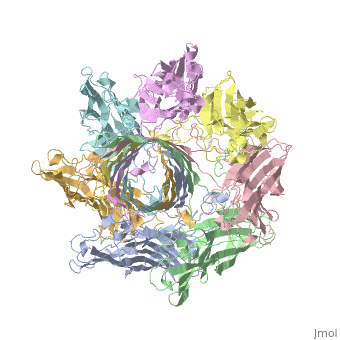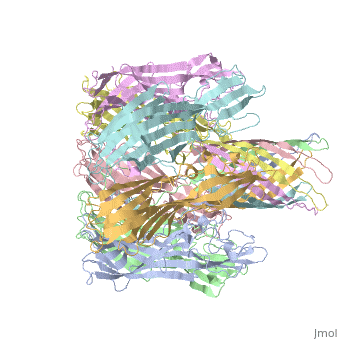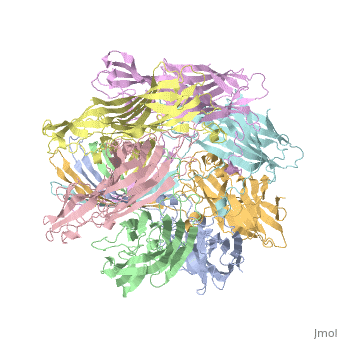Hemolysin: Difference between revisions
Michal Harel (talk | contribs) No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| Line 10: | Line 10: | ||
== Relevance == | == Relevance == | ||
HL acts as a virulence factor in the pathogenesis of invasive infections<ref>PMID:12564994</ref>. | HL acts as a virulence factor in the pathogenesis of invasive infections<ref>PMID:12564994</ref>. | ||
==3D Printed Physical Model of Hemolysin== | ==3D Printed Physical Model of Hemolysin== | ||
| Line 26: | Line 24: | ||
The [http://cbm.msoe.edu MSOE Center for BioMolecular Modeling] uses 3D printing technology to create physical models of protein and molecular structures, making the invisible molecular world more tangible and comprehensible. To view more protein structure models, visit our [http://cbm.msoe.edu/educationalmedia/modelgallery/ Model Gallery]. | The [http://cbm.msoe.edu MSOE Center for BioMolecular Modeling] uses 3D printing technology to create physical models of protein and molecular structures, making the invisible molecular world more tangible and comprehensible. To view more protein structure models, visit our [http://cbm.msoe.edu/educationalmedia/modelgallery/ Model Gallery]. | ||
== 3D Structures of hemolysin == | |||
[[Hemolysin 3D structures]] | |||
</StructureSection> | |||
== 3D Structures of hemolysin == | |||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | ||
| Line 62: | Line 64: | ||
**[[3o44]] – VcHL residues 161-741 – ''Vibrio cholerae''<br /> | **[[3o44]] – VcHL residues 161-741 – ''Vibrio cholerae''<br /> | ||
**[[1xez]] – VcHL (mutant)<br /> | **[[1xez]] – VcHL (mutant)<br /> | ||
**[[6jkz]], [[6jl0]] - VvHL – ''Vibrio vulnificus''<br /> | |||
**[[6jl1]], [[6jl2]] - VvHL (mutant)<br /> | |||
**[[3a57]] – HL 2 – ''Vibrio parahaemolyticus''<br /> | **[[3a57]] – HL 2 – ''Vibrio parahaemolyticus''<br /> | ||
**[[3hvn]] – HL (mutant) – ''Streptococcus suis''<br /> | **[[3hvn]] – HL (mutant) – ''Streptococcus suis''<br /> | ||
| Line 68: | Line 72: | ||
**[[1mt0]] – EcHL B ATP-binding domain – ''Escherichia coli''<br /> | **[[1mt0]] – EcHL B ATP-binding domain – ''Escherichia coli''<br /> | ||
**[[5c21]], [[5c22]] - EcHL D residues 57-333 <br /> | **[[5c21]], [[5c22]] - EcHL D residues 57-333 <br /> | ||
** [[6mru]], [[6mrw]], [[6mrt]] – EcHL E – Cryo EM<br /> | |||
**[[2wcd]] – EcHL E residues 2-303 – ''Escherichia coli''<br /> | **[[2wcd]] – EcHL E residues 2-303 – ''Escherichia coli''<br /> | ||
**[[1qoy]], [[4pho]], [[4phq]] - EcHL E (mutant)<br /> | **[[1qoy]], [[4pho]], [[4phq]] - EcHL E (mutant)<br /> | ||
| Line 73: | Line 78: | ||
**[[2r2z]] – HL residues 346-435 – ''Enterococcus faecalis''<br /> | **[[2r2z]] – HL residues 346-435 – ''Enterococcus faecalis''<br /> | ||
**[[4wx3]], [[4wx5]] - HL – ''Grimontia hollisae''<br /> | **[[4wx3]], [[4wx5]] - HL – ''Grimontia hollisae''<br /> | ||
**[[6d53]], [[6d5z]] - HL C-terminal – ''Bacillus cereus'' - NMR<br /> | |||
*Alpha-toxin | *Alpha-toxin | ||
Revision as of 10:25, 25 July 2019
FunctionHemolysin (HL) is exotoxin from bacteria which causes lysis of red blood cells[1]. Hemolysin from the bacterium Clostridium are called alpha-toxin (AT). AT is a zinc metalloenzyme and binds to the membrane in the presence of calcium. It acts as a phospholipase C. See details for α-hemolysin in Pore forming toxin, α-hemolysin. See details of hemolysin E in Molecular Playground/ClyA. For toxins in Proteopdia see Toxins. RelevanceHL acts as a virulence factor in the pathogenesis of invasive infections[2]. 3D Printed Physical Model of HemolysinShown below is a 3D printed physical model of Hemolysin. The model is shown in alpha carbon backbone format with each chain colored uniquely. The MSOE Center for BioMolecular Modeling The MSOE Center for BioMolecular Modeling uses 3D printing technology to create physical models of protein and molecular structures, making the invisible molecular world more tangible and comprehensible. To view more protein structure models, visit our Model Gallery. 3D Structures of hemolysin
|
| ||||||||||
3D Structures of hemolysin3D Structures of hemolysin
Updated on 25-July-2019
A full page in Proteopedia exploring 7ahl is found here.
- β-hemolysin
- γ-hemolysin
- δ-hemolysin
- 2kam – SaHL-δ - NMR
- Hemolysin
- 3o44 – VcHL residues 161-741 – Vibrio cholerae
- 1xez – VcHL (mutant)
- 6jkz, 6jl0 - VvHL – Vibrio vulnificus
- 6jl1, 6jl2 - VvHL (mutant)
- 3a57 – HL 2 – Vibrio parahaemolyticus
- 3hvn – HL (mutant) – Streptococcus suis
- 3fy3, 5keh, 5kf3, 4w8q, 5kdk – PmHL A residues 30-265 – Proteus mirabilis
- 5sz8, 5kkd, 4w8r, 4w8s, 4w8t - PmHL A residues 30-234 (mutant)
- 1mt0 – EcHL B ATP-binding domain – Escherichia coli
- 5c21, 5c22 - EcHL D residues 57-333
- 6mru, 6mrw, 6mrt – EcHL E – Cryo EM
- 2wcd – EcHL E residues 2-303 – Escherichia coli
- 1qoy, 4pho, 4phq - EcHL E (mutant)
- 2oai, 2r8d – HL corc_hlyc domain – Xylella fastidiosa
- 2r2z – HL residues 346-435 – Enterococcus faecalis
- 4wx3, 4wx5 - HL – Grimontia hollisae
- 6d53, 6d5z - HL C-terminal – Bacillus cereus - NMR
- 3o44 – VcHL residues 161-741 – Vibrio cholerae
- Alpha-toxin
ReferencesReferences
- ↑ Mestre MB, Fader CM, Sola C, Colombo MI. Alpha-hemolysin is required for the activation of the autophagic pathway in Staphylococcus aureus-infected cells. Autophagy. 2010 Jan;6(1):110-25. PMID:20110774
- ↑ Nizet V. Streptococcal beta-hemolysins: genetics and role in disease pathogenesis. Trends Microbiol. 2002 Dec;10(12):575-80. PMID:12564994