NAC transcription factor: Difference between revisions
No edit summary |
No edit summary |
||
| Line 16: | Line 16: | ||
Additionally, the NAC domain also modulates protein binding that may determine fate and function of the NAC protein <ref>http://www.ibt.unam.mx/computo/pdfs/ubiquita/sinat5.pdf</ref> <ref>http://www.biochemj.org/bj/371/0097/3710097.pdf</ref> <ref name="plantc">http://www.plantcell.org/content/22/4/1249.full.pdf+html</ref>. Especially for VNDs, the VNI can directly interact with VND7, and as such, VND7 can directly interact with VND1-5 <ref name="plantc">http://www.plantcell.org/content/22/4/1249.full.pdf+html</ref> <ref name="online">http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2011.04514.x/pdf</ref> Such contacts may also be crucial for plant–pathogen interaction or stress tolerance <ref>http://www.springerlink.com/content/p82h815356615752/fulltext.pdf</ref> <ref>http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2006.02932.x/pdf</ref>. The D subunit of some NAC domains contains a highly hydrophobic negative regulatory domain which acts to suppress transcriptional activity <ref>http://www.springerlink.com/content/x3t8826465j44p32/fulltext.pdf</ref> . Many transcription factor family including Dof, WRKY, and APETALA, can be suppressed. Based on my alignment analyses, most of VNDs in Arabidopsis and poplar have this domain, but the function of this domain for VNDs remain elusive. The hydrophobicity associated with 'LVFY' residues or some structual interference with DNA-binding or nuclear transport in this region may be responsible for such repression. Thanks to the prescence of this domain, the positively charged Lys79, the exposed side chain of Arg85, and the hydrogen bond network of Arg 88 may mediate DNA binding activity <ref>http://www.springerlink.com/content/x3t8826465j44p32/fulltext.pdf</ref> <ref>http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2011.04687.x/pdf</ref>. Furthermore, recent protein structure analyses have shown that NAC domain can change in conformation when binds with DNA <ref>http://www.biochemj.org/bj/imps/pdf/BJ20111742.pdf</ref>. | Additionally, the NAC domain also modulates protein binding that may determine fate and function of the NAC protein <ref>http://www.ibt.unam.mx/computo/pdfs/ubiquita/sinat5.pdf</ref> <ref>http://www.biochemj.org/bj/371/0097/3710097.pdf</ref> <ref name="plantc">http://www.plantcell.org/content/22/4/1249.full.pdf+html</ref>. Especially for VNDs, the VNI can directly interact with VND7, and as such, VND7 can directly interact with VND1-5 <ref name="plantc">http://www.plantcell.org/content/22/4/1249.full.pdf+html</ref> <ref name="online">http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2011.04514.x/pdf</ref> Such contacts may also be crucial for plant–pathogen interaction or stress tolerance <ref>http://www.springerlink.com/content/p82h815356615752/fulltext.pdf</ref> <ref>http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2006.02932.x/pdf</ref>. The D subunit of some NAC domains contains a highly hydrophobic negative regulatory domain which acts to suppress transcriptional activity <ref>http://www.springerlink.com/content/x3t8826465j44p32/fulltext.pdf</ref> . Many transcription factor family including Dof, WRKY, and APETALA, can be suppressed. Based on my alignment analyses, most of VNDs in Arabidopsis and poplar have this domain, but the function of this domain for VNDs remain elusive. The hydrophobicity associated with 'LVFY' residues or some structual interference with DNA-binding or nuclear transport in this region may be responsible for such repression. Thanks to the prescence of this domain, the positively charged Lys79, the exposed side chain of Arg85, and the hydrogen bond network of Arg 88 may mediate DNA binding activity <ref>http://www.springerlink.com/content/x3t8826465j44p32/fulltext.pdf</ref> <ref>http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2011.04687.x/pdf</ref>. Furthermore, recent protein structure analyses have shown that NAC domain can change in conformation when binds with DNA <ref>http://www.biochemj.org/bj/imps/pdf/BJ20111742.pdf</ref>. | ||
*<scene name='48/486354/Cv/ | *<scene name='48/486354/Cv/7'>DNA binding domain</scene>. | ||
*<scene name='48/486354/Cv/ | *<scene name='48/486354/Cv/8'>Dimerization domain</scene>. | ||
*<scene name='48/486354/Cv/ | *<scene name='48/486354/Cv/9'>Salt bridges</scene>. | ||
*<scene name='48/486354/Cv/ | *<scene name='48/486354/Cv/10'>Au coordination sites</scene>. Water molecules shown as red spheres. | ||
== Diverged C-terminal domain == | == Diverged C-terminal domain == | ||
Revision as of 13:03, 18 July 2019
FunctionVascular-related NAC-domain transcription factor (VND) is one group of the largest plant-specific transcription factor NAC family. The VND1-VND7 were orginally isolated as genes for which expression levels are elevated during transdifferentiation into trachery elements, in a induction system using Arabidopsis suspension cells [1].In the past several years, VNDs have been intensively investigated in different species and shown to be important switches of the biosynthesis of secondary cell walls that provide textiles, timber, and potentially second-generation bio-fuels for human use[2][3]. VNDs are grouped in NAC-c subfamily[4]. Typically, the proteins in this subfamily share a well conserved N-terminal NAC domain (-150 amino acid;aa) and a diversified C-terminal transcription regulatory region [5] [6]. The N-terminal NAC domain is usually responsible for DNA binding and dimerization, and the C-terminal region function in transcription activation , repression and protein binding. X-ray crystallography have exhibited the structure of conserved NAC domains when they form dimer and bind with DNA. However, due to the diversified sequence of C-terminal region, no structure analyses haven't been conducted in the region. Conserved NAC domain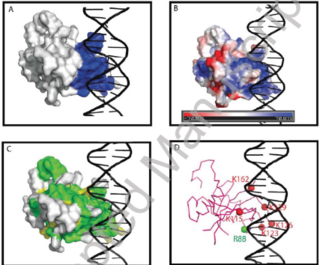 The DNA binding activity of NAC proteins is restricted into NAC domain which was divided into five subdomains A-E. The highly conserved positively charged subdomains C and D bind to DNA, whereas subdomain A may be involved in the formation of a functional dimer. X-ray crystallography[1] have exhibited the presence of a novel transcription factor fold consisting of a twirled antiparallel β-sheet (β 1-6/7) which is used for DNA binding,located between an N-terminal helix and a short helix [7] [8]. Most importantly, Val119-Ser183, lys123 and lys126, along with Lys79, Arg85,and Arg 88 were identified as biochemically crucial for DNA binding. Arg88 is conserved in all NAC proteins but Lys79 and Arg85 could be exchangable but exert different DNA binding affinity [9]. The NAC domain-fold also modulates dimerization through Leu14–Thr23 and Glu26–Tyr31 residues, which form a short antiparallel b-sheet at the dimer interface stabilized by salt bridges formed by Arg19 and Glu26 [6] [7] . This domain also contains mono or bipartite nuclear localization signals with the lysine residues in subdomain D playing crucial roles for nuclear shuttling [3] [10]. Additionally, the NAC domain also modulates protein binding that may determine fate and function of the NAC protein [11] [12] [13]. Especially for VNDs, the VNI can directly interact with VND7, and as such, VND7 can directly interact with VND1-5 [13] [14] Such contacts may also be crucial for plant–pathogen interaction or stress tolerance [15] [16]. The D subunit of some NAC domains contains a highly hydrophobic negative regulatory domain which acts to suppress transcriptional activity [17] . Many transcription factor family including Dof, WRKY, and APETALA, can be suppressed. Based on my alignment analyses, most of VNDs in Arabidopsis and poplar have this domain, but the function of this domain for VNDs remain elusive. The hydrophobicity associated with 'LVFY' residues or some structual interference with DNA-binding or nuclear transport in this region may be responsible for such repression. Thanks to the prescence of this domain, the positively charged Lys79, the exposed side chain of Arg85, and the hydrogen bond network of Arg 88 may mediate DNA binding activity [18] [19]. Furthermore, recent protein structure analyses have shown that NAC domain can change in conformation when binds with DNA [20].
Diverged C-terminal domainThe transcription regulatory region, generally lying at the highly diverged C-terminal, can either activate [21] [22] [23] or repress transcription [13] [24] [25]. Recently, the C-terminal of a novel NAC domain protein VNI have been shown to both activate and repress transcription [26]. More interestingly, VNI2 transcriptional repression motif can be transformed into the transcription activation domain under high salt conditions. It is therefore likely that the C-terminal domain isn’t only complex in sequence, but confer the multiple functions. Based on the sequence analyses, the transcription regulatory region has several group specific motifs that are rich in repeats of serine–threonine, proline–glutamine, or acidic residues, for example, the transcription regulatory region of rice NAC proteins was found to contain ten C-terminal motifs [27]. Another comprehensive study has revealed that these motifs are conserved for a given subgroup of NAC subfamilies but varies across the different subfamilies[2]. Thus, this region imparts variation to individual functions of NAC proteins. Additionally, because of the excessive low-complexity sequences, transcription regulatory regions have a high degree of intrinsic disorder (ID) and fail to have a single stable three-dimensional structure [28][29]. Such flexibility enables them to interact with different target proteins making them model proteins for systematic analysis of transcription factor functions and structural ID. Some NAC proteins have protein-binding ability in their TRRs [24] [28] [30]. An a-helical transmembrane (TM) motif present in some NAC proteins is responsible for plasma membrane or endoplasmic reticulum membrane anchoring [30]. Up to now, 18 membrane bound NAC proteins have been identified in Arabidopsis, 11 in soybean, seven in maize (Zea mays), six in grape, five each in rice, poplar, switchgrass (Panicum virgatum) and sorghum (Sorghum bicolor), and four in Medicago truncatula [31] [32], which may play important regulatory roles under environmental cues. However, no VNDs and other NAC proteins relating to cell wall were identified to have transmembrane motif. The secondary cell wall biosynthesis switches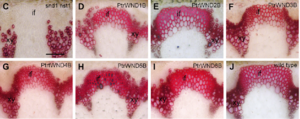 In vascular vessel, VND6 and VND7 control both secondary cell development and programmed cell death of vessels in both root and shoot tissues [1] [33]. The over-expression of VND6 and VND7 can induce ectopic differentiation of two different types of vessel elements: proto-xylem, and meta-xylem vessels. Reversely, the functional repression of VND6 and VND7 can inhibit vessel element formation. Additionally, the excellent works finished by Ye lab showed that the Arabidopsis VND6 and VND7 can complement the NST1NST3 double mutant phenotype, indicating that VNDs share the conserved functions with other secondary cell wall regulators [34]. Then they found that the poplar VNDs can complement the Arabidopsis cell wall development defective mutant NST1NST3, suggesting that the conserved function of VNDs among different species [35]. Recently, the downstream genes of VND6 and VND7 were identified by the excellent works mainly done by Demura lab, Ye lab and Fukuda lab [14] [34] [36]. Both VND6 and VND7 regulates a battery of genes that are common with the downstream of SND1 (secondary cell wall related NAC domain transcription factor), a well known fiber developmental switch. The common downstream genes of VND6, VND7, and SND1 were MYBs transcription factor that have been identified as important regulators of secondary cell wall biosynthesis. However, VND6 and VND7 regulated LBD (Late organ boundery domain) transcription factor that involved in programmed cell death, indicating the important role of VND6 and VND7 in vessel development. The further elucidation of regulatory ways of VNDs not only promote our knowledge in vessel development, but also facilitate the engineering of plant stocks stem from cell wall suitable for biofuel production. |
| ||||||||||
3D Structures of NAC transcription factor3D Structures of NAC transcription factor
Updated on 18-July-2019
3mcb - hNTF NAC domain + nascent polypeptide-associated complex subunit α - human
3lkx - hNTF dimerization domain + nascent polypeptide-associated complex subunit α
3ga1 - hNTF POZ domain
1ut4, 1ut7, 4dul - AtNTF NAC domain - Arabidopsis thaliana
3swp, 3swm - AtNTF NAC domain + DNA
3ulx - NTF NAC domain - rice
ReferenceReference
- 1.Kubo et al (2005) Transcription swtiches for protoxylem and metaxylem vessel formation. Gene Dev.16, 1855-1860.http://genesdev.cshlp.org/content/19/16/1855.full.pdf
- 2.Wang et al (2011) On-Off switches for secondary cell wall biosynthesis. Mol plant. doi:10.1093/mp/ssr098 http://mplant.oxfordjournals.org/content/early/2011/12/01/mp.ssr098.full.pdf+html
- 3.Puranik et al. (2012) NAC proteins: regulation and role in stress tolerance. doi:10.1016/j.tplants.2012.2.14 http://ac.els-cdn.com/S1360138512000428/1-s2.0-S1360138512000428-main.pdf?_tid=9805a6f70ddcc014d8cc606a7deebcd9&acdnat=1336009620_82eaddf71ea7a9126c7de702625e5396
- 4.Shen et al (2009) A bioinformatic analysis of NAC genes for plant cell wall development in relation to lignocellulosic bioenergy production. Bioener.Res. 2,217-232. http://www.springerlink.com/index/qq1584g690243n16.pdf
- 5.Olsen et al (2005) NAC transcription factor : structurally distinct, functionally diverse. Trends Plant Sci. 10,79-87 http://ac.els-cdn.com/S1360138504002961/1-s2.0-S1360138504002961-main.pdf?_tid=6f22ae9d66eced4467706bb9f8c28cc4&acdnat=1336009924_bf0a3f112bc8b062fb6a9916d8a464f4
- 6.Ernst et al (2004)Structure of the conserved domain of ANAC,a member of the NAC family of transcription factors. EMBO J 5,297-303 http://www.nature.com/embor/journal/v5/n3/pdf/7400093.pdf
- 7.Chen et al(2011)A structual view of the conserved domain of rice stress-responsive NAC1. Protein cell 2, 55-63 http://www.springerlink.com/content/t567431215j02gu6/fulltext.pdf
- 8.Puranik et al. (2011) Molecular cloning and characterization of a membrane associated NAC family gene, SiNAC from foxtail millet. Mol. Biotech.49,138-150. http://www.springerlink.com/content/8p88600115713107/fulltext.pdf
- 9.Tran,L.S.P et al. (2009) Molecular characterization of stress-inducable GmNAC genes in soybean.Mol. Genet.Genomics 281.647-664. http://www.springerlink.com/content/r27215773758j405/fulltext.pdf
- 10.Le, D.T. et al. (2011) Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 18, 263–276
http://www.springerlink.com/content/r27215773758j405/fulltext.pdf
- 11.Xie, Q. et al. (2002) SINAT5 promotes ubiquitin-related degradation of NAC1 to attenuate auxin signals. Nature 419, 167–170http://www.ibt.unam.mx/computo/pdfs/ubiquita/sinat5.pdf
- 12. Greve, K. et al. (2003) Interactions between plant RING-H2 and plantspecific NAC (NAM/ATAF1/2/CUC2) proteins: RING-H2 molecular specificity and cellular localization. Biochem. J. 371, 97–108http://www.biochemj.org/bj/371/0097/3710097.pdf
- 13.Yamaguchi, M. et al. (2010) VND-INTERACTING2, a NAC domain transcription factor, negatively regulates xylem vessel formation in
Arabidopsis. Plant Cell 22, 1249–1263 http://www.plantcell.org/content/22/4/1249.full.pdf+html
- 14.Yamaguchi, M. et al. (2011)VASCULAR-RELATED NAC-DOMAIN7 directly regulates the expression of a broad range of genes for xylem vessel formation. Plant Journal 66, 579-90 http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2011.04514.x/pdf
- 15.Xie, Q. et al. (1999) GRAB proteins, novel members of the NAC domain family, isolated by their interaction with a geminivirus protein. Plant Mol. Biol. 39, 647–656 http://www.springerlink.com/content/p82h815356615752/fulltext.pdf
- 16. Tran, L.S.P. et al. (2007) Co-expression of the stress inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 49, 46–63
http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2006.02932.x/pdf
- 17. Hao, Y.J. et al. (2010) Plant NAC-type transcription factor proteins contain a NARD domain for repression of transcriptional activation. Planta 232, 1033–1043 http://www.springerlink.com/content/x3t8826465j44p32/fulltext.pdf
- 18.Hao, Y.J. et al. (2011) Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 68, 302–313http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2011.04687.x/pdf
- 19.Welner, D.H et al.(2012)DNA binding by the plant specific NAC transcription factors in crystal and solution: a firm link to WRKY and GCM transcription factors. Biochem J. doi:10.1092 http://www.biochemj.org/bj/imps/pdf/BJ20111742.pdf
- 20. Tran, L.S.P. et al. (2004) Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought responsive cis-element in the EARLY RESPONSIVE TO
DEHYDRATION STRESS 1 promoter. Plant Cell 16, 2481–2498 http://www.plantcell.org/content/16/9/2481.full.pdf+html
- 21. He, X.J. et al. (2005) AtNAC2, a transcription factor downstream of ethylene and auxin signaling pathways, is involved in salt stress response and lateral root development. Plant J. 44, 903–916
http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2005.02575.x/pdf
- 22. Lu, P.L. et al. (2007) A novel drought-inducible gene, ATAF1, encodes a NAC family protein that negatively regulates the expression of stress-responsive genes in Arabidopsis. Plant Mol. Biol. 63, 289–305 http://www.springerlink.com/content/8101522211447210/fulltext.pdf
- 23. Kim, H.S. et al. (2007) Identification of a calmodulin-binding NAC protein (CBNAC) as a transcriptional repressor in Arabidopsis. J. Biol. Chem. 282, 36292–36302 http://www.jbc.org/content/282/50/36292.full.pdf
- 24. Delessert, C. et al. (2005) The transcription factor ATAF2 represses the expression of pathogenesis-related genes in Arabidopsis. Plant J. 43, 745–757 http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2005.02488.x/pdf
- 25.Yang S.D. et al. (2011) The Arabidopsis NAC Transcription Factor VNI2 Integrates Abscisic Acid Signals into Leaf Senescence via the COR/RD Genes. Plant Cell. 23, 2155–2168 http://www.plantcell.org/content/early/2011/06/13/tpc.111.084913.full.pdf+html
- 26.Fang, Y. et al. (2008) Systematic sequence analysis and identification of tissue-specific or stress-responsive genes of NAC transcription factor family in rice. Mol. Genet. Genomics 280, 535–54 http://www.springerlink.com/content/h437614pg71m7011/fulltext.pdf
- 27.Christiansen, M.W. et al. (2011) Characterization of barley (Hordeum vulgare L.) NAC transcription factors suggests conserved
functions compared to both monocots and dicots. BMC Res. Notes 4,302 http://www.biomedcentral.com/content/pdf/1756-0500-4-302.pdf
- 28 Kjaersgaard, T. et al. (2011) Senescence-associated barley NAC (NAM, ATAF1, 2, CUC) transcription factor interacts with radical-induced cell death 1 through a disordered regulatory domain. J. Biol. Chem. 286, 35418–35429 http://www.jbc.org/content/286/41/35418.full.pdf+html
- 29.Jensen, M.K. et al. (2010) The Arabidopsis thaliana NAC transcription factor family: structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 426, 183–19 http://peer.ccsd.cnrs.fr/docs/00/47/92/44/PDF/PEER_stage2_10.1042%252FBJ20091234.pdf
- 30.Kleinow, T. et al. (2009) NAC domain transcription factor ATAF1 interacts with SNF1-related kinases and silencing of its subfamily
causes severe developmental defects in Arabidopsis. Plant Sci. 177, 360–370 http://www.mpiz-koeln.mpg.de/26442/Kleinow_Plant_J_23_pdf.pdf
- 31.Seo, P.J. et al. (2008) Membrane-bound transcription factors in plants. Trends Plant Sci. 13, 550–556 http://signet.korea.ac.kr/webzine/6th/papers/Trends_in_Plant_Science_200810.pdf
- 32. Kim, S.G. et al. (2010) Genome-scale screening and molecular characterization of membrane-bound transcription factors in Arabidopsis and rice. Genomics 95, 56–6 http://ac.els-cdn.com/S0888754309002122/1-s2.0-S0888754309002122-main.pdf?_tid=f675eda8581767131107a56c803e8434&acdnat=1336012303_7e1cc13e64dad88ebd90823905b9ccfb
- 33. Yamaguchi, M. et al. (2008) Vascular-related NAC-domain 7 is involved in the differentiation of all types of xylem vessels in Arabidopsis roots and shoots. Plant J. 55,652-664 http://onlinelibrary.wiley.com/doi/10.1111/j.1365-313X.2008.03533.x/pdf
- 34. Zhong, R. et al. (2010). Global analysis of direct targets of secondary wall NAC master switches in Arabidopsis. Molecular Plant 3, 1087-1103. http://www.plantbio.uga.edu/~zhye/2010-SWNTargets.pdf
- 35. Zhong, R. et al . (2010) Functional Characterization of Poplar Wood-Associated NAC Domain Transcription Factors. Plant Physiol. 152, 1044-1055 http://www.plantbio.uga.edu/~zhye/2010-PtrWND.pdf
- 36. Ohashi-Ito,K. et al. (2010). Arabidopsis VASCULAR-RELATED NAC-DOMAIN6 Directly Regulates the Genes That Govern Programmed Cell Death and Secondary Wall Formation during Xylem Differentiation. Plant Cell 22,3461–3473 http://www.plantcell.org/content/22/10/3461.full.pdf+html