1eyg: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
==Crystal structure of chymotryptic fragment of E. coli ssb bound to two 35-mer single strand DNAS== | ==Crystal structure of chymotryptic fragment of E. coli ssb bound to two 35-mer single strand DNAS== | ||
<StructureSection load='1eyg' size='340' side='right' caption='[[1eyg]], [[Resolution|resolution]] 2.80Å' scene=''> | <StructureSection load='1eyg' size='340' side='right' caption='[[1eyg]], [[Resolution|resolution]] 2.80Å' scene=''> | ||
== Structural highlights == | == Structural highlights == | ||
<table><tr><td colspan='2'>[[1eyg]] is a 6 chain structure | <table><tr><td colspan='2'>[[1eyg]] is a 6 chain structure. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1EYG OCA]. For a <b>guided tour on the structure components</b> use [http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1EYG FirstGlance]. <br> | ||
</td></tr><tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1kaw|1kaw]]</td></tr> | </td></tr><tr id='related'><td class="sblockLbl"><b>[[Related_structure|Related:]]</b></td><td class="sblockDat">[[1kaw|1kaw]]</td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1eyg FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1eyg OCA], [http://pdbe.org/1eyg PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=1eyg RCSB], [http://www.ebi.ac.uk/pdbsum/1eyg PDBsum]</span></td></tr> | <tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[http://oca.weizmann.ac.il/oca-docs/fgij/fg.htm?mol=1eyg FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1eyg OCA], [http://pdbe.org/1eyg PDBe], [http://www.rcsb.org/pdb/explore.do?structureId=1eyg RCSB], [http://www.ebi.ac.uk/pdbsum/1eyg PDBsum], [http://prosat.h-its.org/prosat/prosatexe?pdbcode=1eyg ProSAT]</span></td></tr> | ||
</table> | </table> | ||
== Function == | == Function == | ||
| Line 16: | Line 17: | ||
<text>to colour the structure by Evolutionary Conservation</text> | <text>to colour the structure by Evolutionary Conservation</text> | ||
</jmolCheckbox> | </jmolCheckbox> | ||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/ | </jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1eyg ConSurf]. | ||
<div style="clear:both"></div> | <div style="clear:both"></div> | ||
<div style="background-color:#fffaf0;"> | <div style="background-color:#fffaf0;"> | ||
| Line 34: | Line 35: | ||
__TOC__ | __TOC__ | ||
</StructureSection> | </StructureSection> | ||
[[Category: Raghunathan, S]] | [[Category: Raghunathan, S]] | ||
[[Category: Waksman, G]] | [[Category: Waksman, G]] | ||
Revision as of 06:55, 6 September 2017
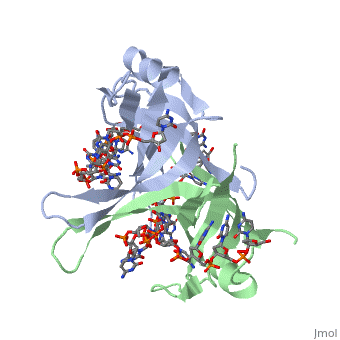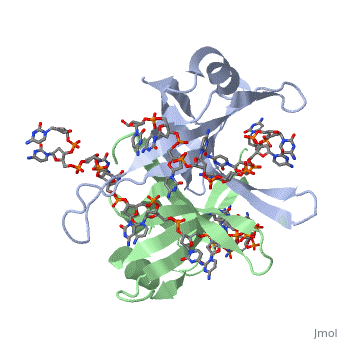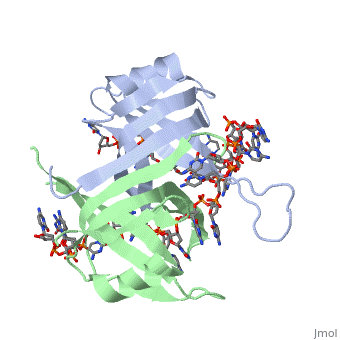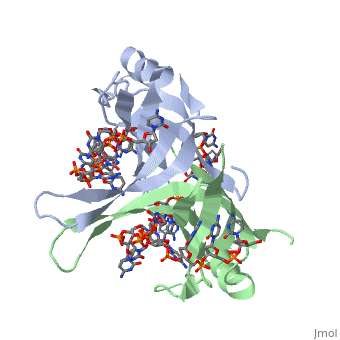
Crystal structure of chymotryptic fragment of E. coli ssb bound to two 35-mer single strand DNASCrystal structure of chymotryptic fragment of E. coli ssb bound to two 35-mer single strand DNAS
Structural highlights
Function[SSB_ECOLI] This protein is essential for replication of the chromosomes and its single-stranded DNA phages. It is also involved in DNA recombination and repair. Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe structure of the homotetrameric DNA binding domain of the single stranded DNA binding protein from Escherichia coli (Eco SSB) bound to two 35-mer single stranded DNAs was determined to a resolution of 2.8 A. This structure describes the vast network of interactions that results in the extensive wrapping of single stranded DNA around the SSB tetramer and suggests a structural basis for its various binding modes. Structure of the DNA binding domain of E. coli SSB bound to ssDNA.,Raghunathan S, Kozlov AG, Lohman TM, Waksman G Nat Struct Biol. 2000 Aug;7(8):648-52. PMID:10932248[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||