Phage integrase: Difference between revisions
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
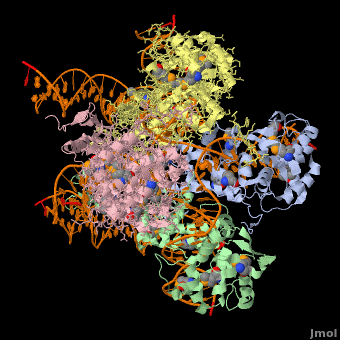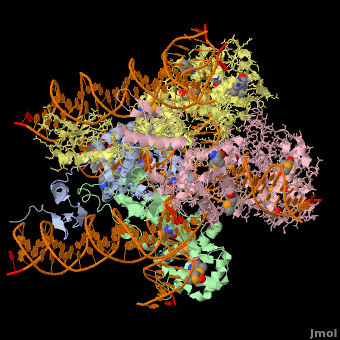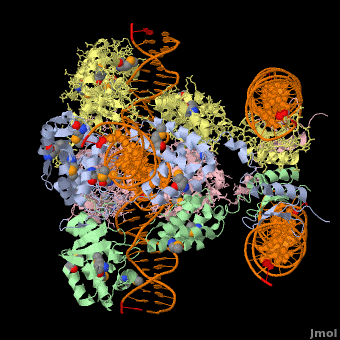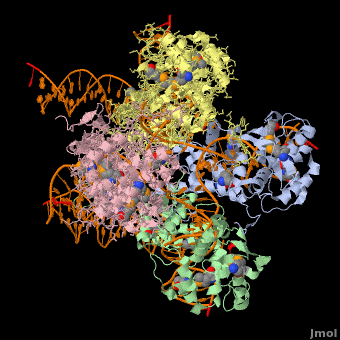
<StructureSection load='1z1g' size='400' side='right' scene=' | <StructureSection load='1z1g' size='400' side='right' scene='44/443479/Cv/2' caption='Se-Met Enterobacteria phage λ integrase:DNA tetramer complex (Holiday Junction) [[1z1g]]' pspeed='8'> | ||
Revision as of 13:48, 29 August 2017
Phage integrase (PIN) mediates recombination between DNA recognition sequences - the phage attachment site attP and the bacterial attachment site attB[1]. The Tyrosine family PIN uses a catalytic tyrosine to mediate strand cleavage while the Serine family PIN uses a serine residue. The High-Pathogenicity Island (HPI) is a genomic island essential for virulence of the family of Enterobacteria. |
| ||||||||||
3D Structures of phage integrase3D Structures of phage integrase
Update June 2011
3nrw – PIN/site-specific recombinase N-terminal – Haloarcula marismortui
3jtz – PIN arm-type binding domain – Yersinia pestis
3ju0 – HPI PIN arm-type binding domain – Pectobacterium atrosepticum
2kkp – PIN SAM-like domain – Moorella thermoacetica – NMR
2kkv – PIN fragment – Salmonella enterica – NMR
2khq - PIN fragment – Staphylococcus saprophyticus – NMR
2wcc – λPIN DBD+ DNA – Enterobacteria phage λ - NMR
1kjk – λPIN N-terminal - NMR
2oxo, 1z19, 1z1b - λPIN core binding domain + DNA
1ae9 - λPIN catalytic core domain (mutant)
1z1g – λPIN + DNA (Holliday junction)
1p7d – λPIN fragment + DNA
3bvp – PIN catalytic core domain – Lactococcus phage TP101-1
1aih - PIN catalytic core domain – Haemophilus phage HP1
2khv, 2kj5 – PIN residues 102-199 – Nitrosospira multiformis - NMR
ReferencesReferences
- ↑ Groth AC, Calos MP. Phage integrases: biology and applications. J Mol Biol. 2004 Jan 16;335(3):667-78. PMID:14687564 doi:http://dx.doi.org/10.1016/S0022283603013561