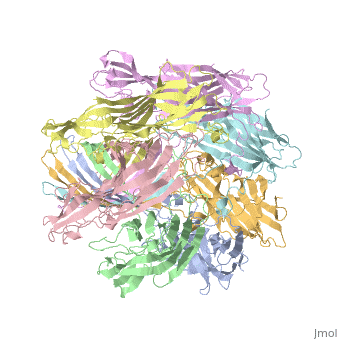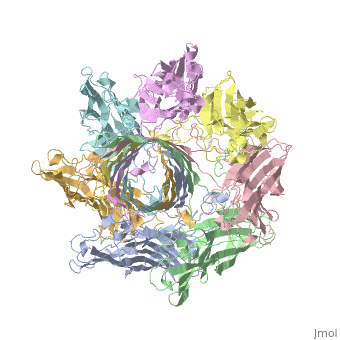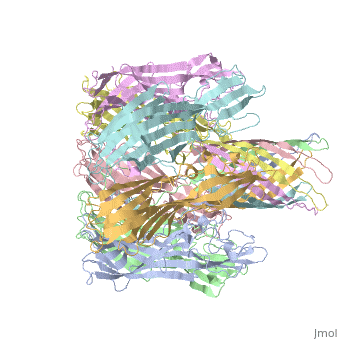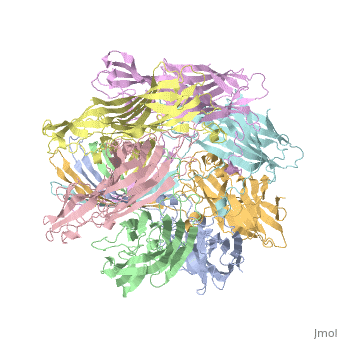Hemolysin: Difference between revisions
Jump to navigation
Jump to search
Michal Harel (talk | contribs) No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
<StructureSection load='7ahl' size='350' side='right' caption='α-hemolysin heptamer (PDB code [[7ahl]]).' scene=''> | <StructureSection load='7ahl' size='350' side='right' caption='α-hemolysin heptamer (PDB code [[7ahl]]).' scene=''> | ||
== Function == | == Function == | ||
| Line 21: | Line 20: | ||
**[[3anz]], [[7ahl]] – SaHL-α – ''Staphylococcus aureus''<br /> | **[[3anz]], [[7ahl]] – SaHL-α – ''Staphylococcus aureus''<br /> | ||
A full page in Proteopedia exploring [[7ahl]] is found [[Pore_forming_toxin,_α-hemolsyin|here]].<br/> | A full page in Proteopedia exploring [[7ahl]] is found [[Pore_forming_toxin,_α-hemolsyin|here]].<br/> | ||
**[[3m2l]], [[3m4d]], [[4p24]] - SaHL-α (mutant)<br /> | **[[3m2l]], [[3m4d]], [[4p24]], [[4yhd]] - SaHL-α (mutant)<br /> | ||
**[[3m3r]], [[3m4e]] - SaHL-α (mutant) + β-cyclodextrin<br /> | **[[3m3r]], [[3m4e]] - SaHL-α (mutant) + β-cyclodextrin<br /> | ||
**[[4idj]], [[4u6v]] - SaHL-α + antibody<br /> | **[[4idj]], [[4u6v]] - SaHL-α + antibody<br /> | ||
| Line 49: | Line 48: | ||
**[[3a57]] – HL 2 – ''Vibrio parahaemolyticus''<br /> | **[[3a57]] – HL 2 – ''Vibrio parahaemolyticus''<br /> | ||
**[[3hvn]] – HL (mutant) – ''Streptococcus suis''<br /> | **[[3hvn]] – HL (mutant) – ''Streptococcus suis''<br /> | ||
**[[3fy3]] – | **[[3fy3]], [[5keh]], [[5kf3]], [[4w8q]] – PmHL A residues 30-265 – ''Proteus mirabilis''<br /> | ||
**[[5sz8]], [[5kkd]], [[4w8r]], [[4w8s]], [[4w8t]] - PmHL A residues 30-234 (mutant)<br /> | |||
**[[1mt0]] – EcHL B ATP-binding domain – ''Escherichia coli''<br /> | |||
**[[5c21]], [[5c22]] - EcHL D residues 57-333 <br /> | |||
**[[2wcd]] – EcHL E residues 2-303 – ''Escherichia coli''<br /> | **[[2wcd]] – EcHL E residues 2-303 – ''Escherichia coli''<br /> | ||
**[[1qoy]], [[4pho]], [[4phq]] - EcHL E (mutant)<br /> | **[[1qoy]], [[4pho]], [[4phq]] - EcHL E (mutant)<br /> | ||
**[[2oai]], [[2r8d]] – HL corc_hlyc domain – ''Xylella fastidiosa''<br /> | **[[2oai]], [[2r8d]] – HL corc_hlyc domain – ''Xylella fastidiosa''<br /> | ||
**[[2r2z]] – HL residues 346-435 – ''Enterococcus faecalis''<br /> | **[[2r2z]] – HL residues 346-435 – ''Enterococcus faecalis''<br /> | ||
**[[4wx3]], [[4wx5]] - HL – ''Grimontia hollisae''<br /> | |||
*Alpha-toxin | *Alpha-toxin | ||
Revision as of 11:16, 12 June 2017
FunctionHemolysin (HL) is exotoxin from bacteria which causes lysis of red blood cells[1]. Hemolysin from the bacterium Clostridium are called alpha-toxin (AT). AT is a zinc metalloenzyme and binds to the membrane in the presence of calcium. It acts as a phospholipase C. See details for α-hemolysin in Pore forming toxin, α-hemolysin. See details of hemolysin E in Molecular Playground/ClyA. For toxins in Proteopdia see Toxins. RelevanceHL acts as a virulence factor in the pathogenesis of invasive infections[2]. |
| ||||||||||
3D Structures of hemolysin3D Structures of hemolysin
Updated on 12-June-2017
A full page in Proteopedia exploring 7ahl is found here.
- β-hemolysin
- γ-hemolysin
- δ-hemolysin
- 2kam – SaHL-δ - NMR
- Hemolysin
- 3o44 – VcHL residues 161-741 – Vibrio cholerae
- 1xez – VcHL (mutant)
- 3a57 – HL 2 – Vibrio parahaemolyticus
- 3hvn – HL (mutant) – Streptococcus suis
- 3fy3, 5keh, 5kf3, 4w8q – PmHL A residues 30-265 – Proteus mirabilis
- 5sz8, 5kkd, 4w8r, 4w8s, 4w8t - PmHL A residues 30-234 (mutant)
- 1mt0 – EcHL B ATP-binding domain – Escherichia coli
- 5c21, 5c22 - EcHL D residues 57-333
- 2wcd – EcHL E residues 2-303 – Escherichia coli
- 1qoy, 4pho, 4phq - EcHL E (mutant)
- 2oai, 2r8d – HL corc_hlyc domain – Xylella fastidiosa
- 2r2z – HL residues 346-435 – Enterococcus faecalis
- 4wx3, 4wx5 - HL – Grimontia hollisae
- 3o44 – VcHL residues 161-741 – Vibrio cholerae
- Alpha-toxin
ReferencesReferences
- ↑ Mestre MB, Fader CM, Sola C, Colombo MI. Alpha-hemolysin is required for the activation of the autophagic pathway in Staphylococcus aureus-infected cells. Autophagy. 2010 Jan;6(1):110-25. PMID:20110774
- ↑ Nizet V. Streptococcal beta-hemolysins: genetics and role in disease pathogenesis. Trends Microbiol. 2002 Dec;10(12):575-80. PMID:12564994