Amyloid precursor protein: Difference between revisions
Michal Harel (talk | contribs) No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
{{STRUCTURE_1mwp| PDB=1mwp | SIZE= | {{STRUCTURE_1mwp| PDB=1mwp | SIZE=350| SCENE= |right| CAPTION=Human amyloid precursor protein heparin-binding domain [[1mwp]] }} | ||
== Function == | == Function == | ||
Revision as of 12:38, 2 December 2015
| |||||||||
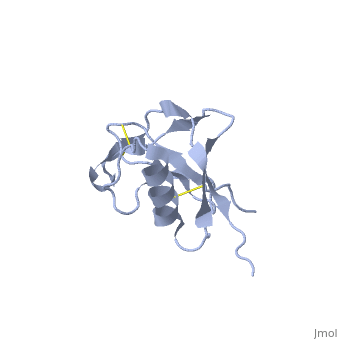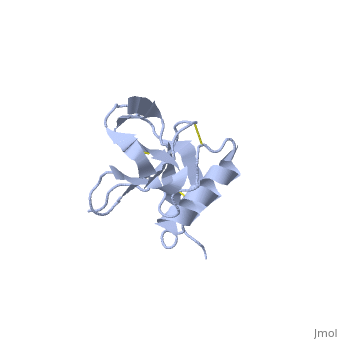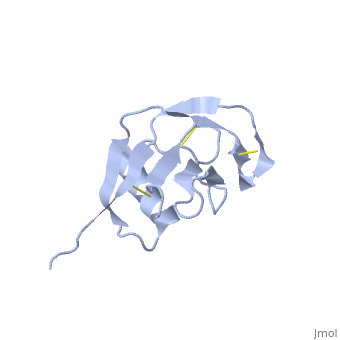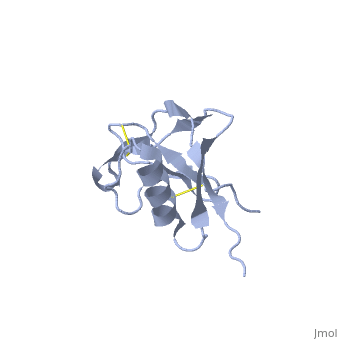
| Human amyloid precursor protein heparin-binding domain 1mwp | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
FunctionFunction
Amyloid precursor protein (APP) is a transmembranal protein which is thought to regulate transcription. APP plays a role in synaptic formation and repair.
DiseaseDisease
APP is cleaved by β-secretase and the resulting N terminal peptide which is ca. 40 amino acid long is called hAPP β-peptide. The aggregation of the hAPP β-peptide is the cause of Alzheimer’s Disease. For detailed discussion see
Structural highlightsStructural highlights
The extracellular region of APP contains several domains named E1 (residues 1-189) and E2 (residues 346-551). The E1 domain contains a growth factor-like or heparin-binding (residues 28-123) and Cu-binding (residues 124-189) subdomains. Additional domains are: Kunitz-type protease inhibitor (residues 287-344) and Zn-binding (residues 672-687). The Z-binding domain is involved in the oligomerizatin of APP.
3D structures of amyloid precursor protein3D structures of amyloid precursor protein
Updated on 02-December-2015