Clp Protease: Difference between revisions
Jump to navigation
Jump to search
Michal Harel (talk | contribs) No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| Line 18: | Line 18: | ||
**[[1tg6]] - CLP – human<br /> | **[[1tg6]] - CLP – human<br /> | ||
**[[2f6i]], [[4gm2]], [[4hnk]] - CLP – ''Plasmodium falciparum''<br /> | **[[2f6i]], [[4gm2]], [[4hnk]] - CLP – ''Plasmodium falciparum''<br /> | ||
**[[2cby]], [[2ce3]], [[2c8t]] - | **[[2cby]], [[2ce3]], [[2c8t]], [[4u0h]] - MtCLP – ''Mycobacterium tuberculosis''<br /> | ||
**[[3fes]] - CLP – ''Clostridium difficile''<br /> | **[[3fes]] - CLP – ''Clostridium difficile''<br /> | ||
**[[3ktg]], [[3kth]], [[3tt6]] – BsCLP – ''Bacillus subtilis''<br /> | **[[3ktg]], [[3kth]], [[3tt6]] – BsCLP – ''Bacillus subtilis''<br /> | ||
| Line 27: | Line 27: | ||
**[[2zl0]] - HpCLP – ''Helicobacter pylori''<br /> | **[[2zl0]] - HpCLP – ''Helicobacter pylori''<br /> | ||
**[[2zl3]] - HpCLP (mutant)<br /> | **[[2zl3]] - HpCLP (mutant)<br /> | ||
**[[4jcq]], [[4jct]] – LmCLP – ''Listeria monocytogenes''<br /> | **[[4jcq]], [[4jct]], [[4ryf]] – LmCLP – ''Listeria monocytogenes''<br /> | ||
**[[4jcr]] - LmCLP (mutant)<br /> | **[[4jcr]] - LmCLP (mutant)<br /> | ||
*''CLP P subunit | *''CLP P subunit complex'' | ||
**[[2fzs]], [[3mt6]] - EcCLP + peptide<br /> | **[[2fzs]], [[3mt6]] - EcCLP + peptide<br /> | ||
| Line 36: | Line 36: | ||
**[[2zl4]] - HpCLP (mutant) + peptide<br /> | **[[2zl4]] - HpCLP (mutant) + peptide<br /> | ||
**[[3kti]], [[3ktj]], [[3ktk]] - BsCLP + peptide<br /> | **[[3kti]], [[3ktj]], [[3ktk]] - BsCLP + peptide<br /> | ||
**[[3tt7]] - BsCLP + DFP | **[[3tt7]] - BsCLP + DFP<br /> | ||
**[[4u0g]] – MtCLP + ADEP + agonist<br /> | |||
*CLP A subunit | *CLP A subunit | ||
Revision as of 13:06, 16 March 2015
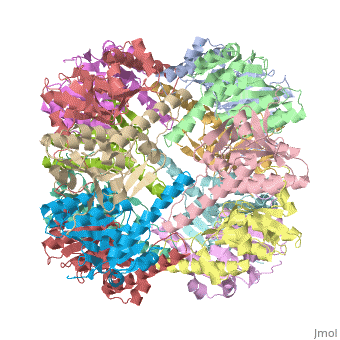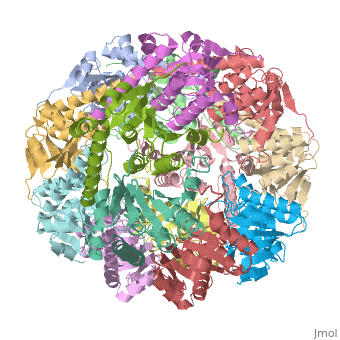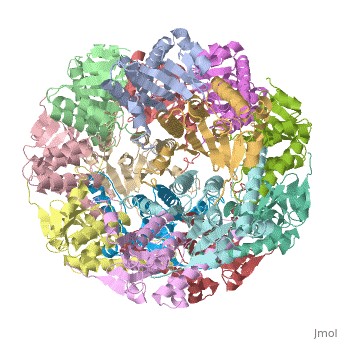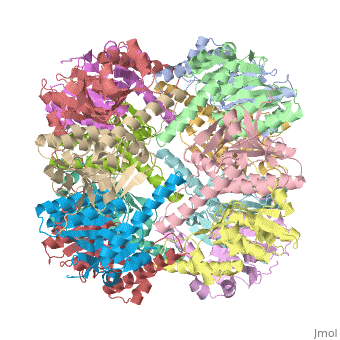
| |||||||||
| E. coli Clp protease, 1tyf | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Activity: | Endopeptidase Clp, with EC number 3.4.21.92 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
Clp protease (CLP) is a serine peptidase which hydrolyzes proteins in the presence of ATP and Mg+2 ion. CLP is found in mitochondria. CLP participates in degradation of misfolded proteins. CLP is a heterodimer containing an ATP-binding regulatory subunit A and catalytic subunit P. For more details see
3D structures of Clp protease3D structures of Clp protease
Updated on 16-March-2015