Glyceraldehyde-3-Phosphate Dehydrogenase: Difference between revisions
Michal Harel (talk | contribs) No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
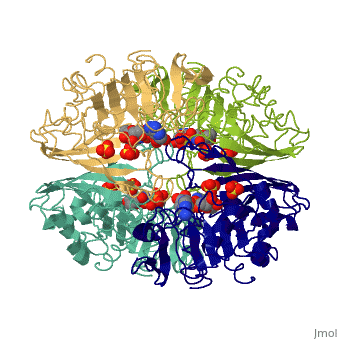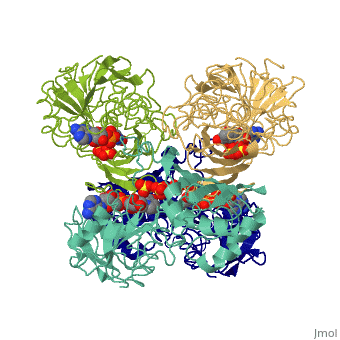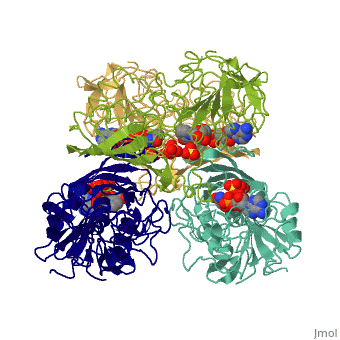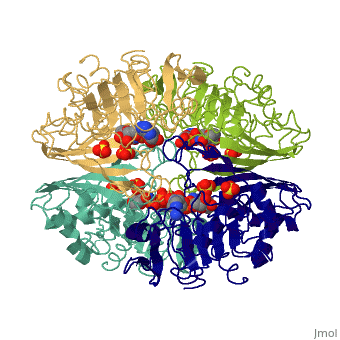
{{STRUCTURE_3gpd| PDB=3gpd | SIZE=400| SCENE= |right|CAPTION=Human glyceraldehyde-3-phosphate dehydrogenase complex with NAD and sulfate, [[3gpd]] }} | {{STRUCTURE_3gpd| PDB=3gpd | SIZE=400| SCENE= |right|CAPTION=Human glyceraldehyde-3-phosphate dehydrogenase complex with NAD and sulfate, [[3gpd]] }} | ||
| Line 39: | Line 38: | ||
===Apo GAPDH=== | ===Apo GAPDH=== | ||
1vsu – CpGAPDH – ''Cryptosporidium parvum''<br /> | [[1vsu]] – CpGAPDH – ''Cryptosporidium parvum''<br /> | ||
1dc5 - EcGAPDH – ''Escherichia coli''<br /> | [[1dc5]] - EcGAPDH – ''Escherichia coli''<br /> | ||
1euh, 1qi6 - SmGAPDH – ''Streptococcus mutans''<br /> | [[1euh]], [[1qi6]] - SmGAPDH – ''Streptococcus mutans''<br /> | ||
2hki – sGAPDH – spinach<br /> | [[2hki]] – sGAPDH – spinach<br /> | ||
3e6a – RiGAPDH ligand binding domain – rice<br /> | [[3e6a]] – RiGAPDH ligand binding domain – rice<br /> | ||
3lc7 - SaGAPDH – ''Staphylococcus aureus''<br /> | [[3lc7]] - SaGAPDH – ''Staphylococcus aureus''<br /> | ||
3pqa – MjGAPDH – ''Methanocaldococcus jannaschii''<br /> | [[3pqa]] – MjGAPDH – ''Methanocaldococcus jannaschii''<br /> | ||
3prl – BhGAPDH – ''Bacillus halodurans''<br /> | [[3prl]] – BhGAPDH – ''Bacillus halodurans''<br /> | ||
===NAD-dependent GAPDH with cofactor NAD=== | ===NAD-dependent GAPDH with cofactor NAD=== | ||
1vsv – CpGAPDH + NAD<br /> | [[1vsv]] – CpGAPDH + NAD<br /> | ||
1gpd – GAPDH + NAD – American lobster<br /> | [[1gpd]] – GAPDH + NAD – American lobster<br /> | ||
3gpd, 1znq, 1u8f - GAPDH + NAD – human<br /> | [[3gpd]], [[1znq]], [[1u8f]] - GAPDH + NAD – human<br /> | ||
2vyn, 2vyv - GAPDH + NAD - rat<br /> | [[2vyn]], [[2vyv]] - GAPDH + NAD - rat<br /> | ||
1gd1, 3cmc – GsGAPDH + NAD – ''Geobacillus stearothermophilus''<br /> | [[1gd1]], [[3cmc]] – GsGAPDH + NAD – ''Geobacillus stearothermophilus''<br /> | ||
1dbv, 3dbv - GsGAPDH (mutant) + NAD<br /> | [[1dbv]], [[3dbv]] - GsGAPDH (mutant) + NAD<br /> | ||
1hdg - GAPDH + NAD – ''Thermotoga maritima''<br /> | [[1hdg]] - GAPDH + NAD – ''Thermotoga maritima''<br /> | ||
1gyp, 1a7k - GAPDH + NAD – ''Leishmania mexicana''<br /> | [[1gyp]], [[1a7k]] - GAPDH + NAD – ''Leishmania mexicana''<br /> | ||
1gyq - LmGAPDH + benzyl-NAD<br /> | [[1gyq]] - LmGAPDH + benzyl-NAD<br /> | ||
1cer, 2g82 - GAPDH + NAD – ''Thermus aquaticus''<br /> | [[1cer]], [[2g82]] - GAPDH + NAD – ''Thermus aquaticus''<br /> | ||
1gad, 1dc6 - EcGAPDH + NAD<br /> | [[1gad]], [[1dc6]] - EcGAPDH + NAD<br /> | ||
1gae - EcGAPDH (mutant) + NAD<br /> | [[1gae]] - EcGAPDH (mutant) + NAD<br /> | ||
2ep7 – GAPDH + NAD – ''Aquifex aeolicus''<br /> | [[2ep7]] – GAPDH + NAD – ''Aquifex aeolicus''<br /> | ||
1nbo - sGAPDH + NAD<br /> | [[1nbo]] - sGAPDH + NAD<br /> | ||
1j0x - GAPDH + NAD – rabbit<br /> | [[1j0x]] - GAPDH + NAD – rabbit<br /> | ||
2b4r, 1ywg - PfGAPDH + NAD – ''Plasmodium falciparum''<br /> | [[2b4r]], [[1ywg]] - PfGAPDH + NAD – ''Plasmodium falciparum''<br /> | ||
2b4t - PfGAPDH (mutant) + NAD<br /> | [[2b4t]] - PfGAPDH (mutant) + NAD<br /> | ||
2czc - GAPDH + NAD – ''Pyrococcus horikoshii''<br /> | [[2czc]] - GAPDH + NAD – ''Pyrococcus horikoshii''<br /> | ||
2i5p - GAPDH + NAD – ''Kluyveromyces marxianus''<br /> | [[2i5p]] - GAPDH + NAD – ''Kluyveromyces marxianus''<br /> | ||
3gnq - GAPDH + NAD – ''Burkholderia pseudomallei''<br /> | [[3gnq]] - GAPDH + NAD – ''Burkholderia pseudomallei''<br /> | ||
3hja - GAPDH + NAD – ''Borrelia burgdorferi''<br /> | [[3hja]] - GAPDH + NAD – ''Borrelia burgdorferi''<br /> | ||
3e5r - RiGAPDH + NAD<br /> | [[3e5r]] - RiGAPDH + NAD<br /> | ||
2x0n – GAPDH + NAD – ''Trypanosoma brucei'' – Laue<br /> | [[2x0n]] – GAPDH + NAD – ''Trypanosoma brucei'' – Laue<br /> | ||
3k2b - GAPDH + NAD – ''Arabidopsis thaliana''<br /> | [[3k2b]] - GAPDH + NAD – ''Arabidopsis thaliana''<br /> | ||
3hq4, 3k9q, 3ksd, 3lc1 - SaGAPDH (mutant) + NAD<br /> | [[3hq4]], [[3k9q]], [[3ksd]], [[3lc1]] - SaGAPDH (mutant) + NAD<br /> | ||
3lvf - SaGAPDH + NAD<br /> | [[3lvf]] - SaGAPDH + NAD<br /> | ||
3pfw - hGAPDH + NAD – human<br /> | [[3pfw]] - hGAPDH + NAD – human<br /> | ||
3sth - GAPDH + NAD – ''Toxoplasma gondii''<br /> | [[3sth]] - GAPDH + NAD – ''Toxoplasma gondii''<br /> | ||
3pym - GAPDH + NAD – yeast<br /> | [[3pym]] - GAPDH + NAD – yeast<br /> | ||
===NAD-dependent GAPDH ternary complexes containing glyceraldehyde-3-phosphate (G3P)=== | ===NAD-dependent GAPDH ternary complexes containing glyceraldehyde-3-phosphate (G3P)=== | ||
3cif - CpGAPDH (mutant) + NAD + G3P<br /> | [[3cif]] - CpGAPDH (mutant) + NAD + G3P<br /> | ||
3ksz, 3l4s - SaGAPDH (mutant) + NAD + G3P<br /> | [[3ksz]], [[3l4s]] - SaGAPDH (mutant) + NAD + G3P<br /> | ||
===Other NAD-dependent GAPDH ternary complexes=== | ===Other NAD-dependent GAPDH ternary complexes=== | ||
3dmt, 3ids - TcGAPDH + NAD + inhibitor<br /> | [[3dmt]], [[3ids]] - TcGAPDH + NAD + inhibitor<br /> | ||
3h9e - hGAPDH + NAD + phosphate<br /> | [[3h9e]] - hGAPDH + NAD + phosphate<br /> | ||
3k73, 3l6o - SaGAPDH + NAD + phosphate<br /> | [[3k73]], [[3l6o]] - SaGAPDH + NAD + phosphate<br /> | ||
1uxt - TtGAPDH (mutant) + NAD + G1P<br /> | [[1uxt]] - TtGAPDH (mutant) + NAD + G1P<br /> | ||
3b1j, 3b1k - GAPDH + NAD + CP12 – ''Synechococcus elongatus''<br /> | [[3b1j]], [[3b1k]] - GAPDH + NAD + CP12 – ''Synechococcus elongatus''<br /> | ||
===NADP-dependent GAPDH with cofactor NADP=== | ===NADP-dependent GAPDH with cofactor NADP=== | ||
2dbv, 4dbv - GsGAPDH (mutant) + NADP<br /> | [[2dbv]], [[4dbv]] - GsGAPDH (mutant) + NADP<br /> | ||
2yyy - MjGAPDH + NADP<br /> | [[2yyy]] - MjGAPDH + NADP<br /> | ||
2euh - SmGAPDH + NADP<br /> | [[2euh]] - SmGAPDH + NADP<br /> | ||
2id2 - SmGAPDH (mutant) + NADP<br /> | [[2id2]] - SmGAPDH (mutant) + NADP<br /> | ||
1cf2 - GAPDH + NADP – ''Methanothermus fervidus''<br /> | [[1cf2]] - GAPDH + NADP – ''Methanothermus fervidus''<br /> | ||
1jn0, 1rm4, 2pkq, 2pkr - sGAPDH + NADP<br /> | [[1jn0]], [[1rm4]], [[2pkq]], [[2pkr]] - sGAPDH + NADP<br /> | ||
1rm3, 1rm5 - sGAPDH (mutant) + NADP<br /> | [[1rm3]], [[1rm5]] - sGAPDH (mutant) + NADP<br /> | ||
1ky8 - TtGAPDH + NADP – ''Thermoproteus tenax''<br /> | [[1ky8]] - TtGAPDH + NADP – ''Thermoproteus tenax''<br /> | ||
3rhh - BhGAPDH + NADP<br /> | [[3rhh]] - BhGAPDH + NADP<br /> | ||
===NADP-dependent GAPDH ternary complexes containing glyceraldehyde-3-phosphate (G3P)=== | ===NADP-dependent GAPDH ternary complexes containing glyceraldehyde-3-phosphate (G3P)=== | ||
1qi1 - SmGAPDH + NADP + G3P<br /> | [[1qi1]] - SmGAPDH + NADP + G3P<br /> | ||
2esd, 2qe0 - SmGAPDH (mutant) + NADP + G3P<br /> | [[2esd]], [[2qe0]] - SmGAPDH (mutant) + NADP + G3P<br /> | ||
3lc2 – SaGAPDH-thioacyl + G3P<br /> | [[3lc2]] – SaGAPDH-thioacyl + G3P<br /> | ||
1uxu - TtGAPDH (mutant) + NADP + AMP + G3P<br /> | [[1uxu]] - TtGAPDH (mutant) + NADP + AMP + G3P<br /> | ||
===Other NADP-dependent GAPDH ternary complexes=== | ===Other NADP-dependent GAPDH ternary complexes=== | ||
1k3t - TcGAPDH + coumarin derivative – ''Trypanosoma cruzi''<br /> | [[1k3t]] - TcGAPDH + coumarin derivative – ''Trypanosoma cruzi''<br /> | ||
1uxv - TtGAPDH + NADP + AMP<br /> | [[1uxv]] - TtGAPDH + NADP + AMP<br /> | ||
1uxp - TtGAPDH (mutant) + NADP + ADP<br /> | [[1uxp]] - TtGAPDH (mutant) + NADP + ADP<br /> | ||
1uxq - TtGAPDH (mutant) + NADP + G1P<br /> | [[1uxq]] - TtGAPDH (mutant) + NADP + G1P<br /> | ||
1uxr - TtGAPDH (mutant) + NADP + F6P<br /> | [[1uxr]] - TtGAPDH (mutant) + NADP + F6P<br /> | ||
==References== | ==References== | ||
Revision as of 12:10, 24 August 2014
| |||||||||
| Human glyceraldehyde-3-phosphate dehydrogenase complex with NAD and sulfate, 3gpd | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| Activity: | Glyceraldehyde-3-phosphate dehydrogenase (phosphorylating), with EC number 1.2.1.12 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, PDBsum, RCSB | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
FunctionFunction
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a very important enzyme in the production of energy and in photosynthesis. In the production of energy this enzyme catalyzes the sixth step in the process of breaking down glucose, also known as glycolysis which occurs in organisms of all phyla. The sixth step consists of of the oxidation of GAP by NAD and an inorganic phosphate to yield 1,3 bisphosphoglycerate. In photosynthesis, which is carried out by plants and algae, this enzyme uses NADPH in the reverse reaction in a step in the Calvin Cycle, which fixes gaseous CO2 into carbohydrate. Though these are its main functions, GAPDH has been shown to perform other functions including transcription activation, initiation of apoptosis, and ER to Golgi apparatus vesicle transportation [1]. However, this page will focus on GAPDH’s role in glycolysis. See 2pkq for the plant Calvin Cycle enzyme.
StructureStructure
GAPDH most commonly exists as what looks to be a dimer. Interesting though, the two monomers of the enzyme are not exactly the same. While one side consists only of parallel and antiparallel beta-sheets, the other monomer is made up of both . Though each monomer does not have to exact same sequence, each does contain replicate active sites and function. This is consistent with the following SCOP information:
Class: Alpha and beta proteins (a/b) Fold: NAD(P)-binding Rossmann-fold domains Superfamily: NAD(P)-binding Rossmann-fold domains Family: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminus domain Protein: Glyceraldehyde-3-phosphate dehydrogenase Species: Human
The specific reaction that GAPDH catalyzes is shown below:
GAP + NAD+ + Pi +GAPDH <==> 1,3-bisphosphoglycerate + NADH + H
RegulationRegulation
GAPDH is not a highly regulated step with in the glycolytic pathway because of relatively low energy transitions that occur between steps five and nine. However, there are a few items that must be present for the reaction to proceed. GAPDH is inactivated by the alkylation of iodoacetate, in which the Iodine inhibits the active site by binding to the Sulfur present in the cystine residue. If the active site is blocked by by an inhibitor such as Iodine then the reaction will stop. Also NAD oxidizes the GAP, so the availability of hydrogen and inorganic phosphates also could effect the rates of the reaction. The reason this step is not highly regulated is because Iodine is not present in the blood stream and the absence of both hydrogen and inorganic phosphates will cause the reaction to yield before this step is met.
Reaction MechanismReaction Mechanism
The mechanism of the glycolysis reaction is fairly straight forward. After the aldehyde enters the (highlighted in green), the sulfhydryl group from attacks the nucleophilic carbon to form a thiohemiacetal. This intermediate undergoes oxidation due to a hydride transfer to a nearby NAD+ forming a thioester. From here, a phosphate group enters and attacks the same carbonyl while at the same time it is separated from the cystine by the protonated group. This produces the desired 1,3-bisphosphoglycerate. Though cysteine-151 and histidine-178 are direct contributers to the catalytic process, other residues also influence the activity of this enzyme indirectly. are two such residues that contribute to the binding of the reactants rather than the catalytic mechanism. Regulation of GAPDH occurs through its coupling with the PGK reaction. This coupling is needed due to the slightly positive delta G of the glycolysis. The larger negative delta G of the PGK reaction results in the following overall net reaction with a delta G of -12.1 kJ/mol:
GAP + Pi + NAD+ + ADP ==> 3PG + NADH + ATP
Additional ResourcesAdditional Resources
For additional information, see: Carbohydrate Metabolism
3D structures of glyceraldehyde-3-phosphate dehydrogenase3D structures of glyceraldehyde-3-phosphate dehydrogenase
Updated on 24-August-2014
Apo GAPDHApo GAPDH
1vsu – CpGAPDH – Cryptosporidium parvum
1dc5 - EcGAPDH – Escherichia coli
1euh, 1qi6 - SmGAPDH – Streptococcus mutans
2hki – sGAPDH – spinach
3e6a – RiGAPDH ligand binding domain – rice
3lc7 - SaGAPDH – Staphylococcus aureus
3pqa – MjGAPDH – Methanocaldococcus jannaschii
3prl – BhGAPDH – Bacillus halodurans
NAD-dependent GAPDH with cofactor NADNAD-dependent GAPDH with cofactor NAD
1vsv – CpGAPDH + NAD
1gpd – GAPDH + NAD – American lobster
3gpd, 1znq, 1u8f - GAPDH + NAD – human
2vyn, 2vyv - GAPDH + NAD - rat
1gd1, 3cmc – GsGAPDH + NAD – Geobacillus stearothermophilus
1dbv, 3dbv - GsGAPDH (mutant) + NAD
1hdg - GAPDH + NAD – Thermotoga maritima
1gyp, 1a7k - GAPDH + NAD – Leishmania mexicana
1gyq - LmGAPDH + benzyl-NAD
1cer, 2g82 - GAPDH + NAD – Thermus aquaticus
1gad, 1dc6 - EcGAPDH + NAD
1gae - EcGAPDH (mutant) + NAD
2ep7 – GAPDH + NAD – Aquifex aeolicus
1nbo - sGAPDH + NAD
1j0x - GAPDH + NAD – rabbit
2b4r, 1ywg - PfGAPDH + NAD – Plasmodium falciparum
2b4t - PfGAPDH (mutant) + NAD
2czc - GAPDH + NAD – Pyrococcus horikoshii
2i5p - GAPDH + NAD – Kluyveromyces marxianus
3gnq - GAPDH + NAD – Burkholderia pseudomallei
3hja - GAPDH + NAD – Borrelia burgdorferi
3e5r - RiGAPDH + NAD
2x0n – GAPDH + NAD – Trypanosoma brucei – Laue
3k2b - GAPDH + NAD – Arabidopsis thaliana
3hq4, 3k9q, 3ksd, 3lc1 - SaGAPDH (mutant) + NAD
3lvf - SaGAPDH + NAD
3pfw - hGAPDH + NAD – human
3sth - GAPDH + NAD – Toxoplasma gondii
3pym - GAPDH + NAD – yeast
NAD-dependent GAPDH ternary complexes containing glyceraldehyde-3-phosphate (G3P)NAD-dependent GAPDH ternary complexes containing glyceraldehyde-3-phosphate (G3P)
3cif - CpGAPDH (mutant) + NAD + G3P
3ksz, 3l4s - SaGAPDH (mutant) + NAD + G3P
Other NAD-dependent GAPDH ternary complexesOther NAD-dependent GAPDH ternary complexes
3dmt, 3ids - TcGAPDH + NAD + inhibitor
3h9e - hGAPDH + NAD + phosphate
3k73, 3l6o - SaGAPDH + NAD + phosphate
1uxt - TtGAPDH (mutant) + NAD + G1P
3b1j, 3b1k - GAPDH + NAD + CP12 – Synechococcus elongatus
NADP-dependent GAPDH with cofactor NADPNADP-dependent GAPDH with cofactor NADP
2dbv, 4dbv - GsGAPDH (mutant) + NADP
2yyy - MjGAPDH + NADP
2euh - SmGAPDH + NADP
2id2 - SmGAPDH (mutant) + NADP
1cf2 - GAPDH + NADP – Methanothermus fervidus
1jn0, 1rm4, 2pkq, 2pkr - sGAPDH + NADP
1rm3, 1rm5 - sGAPDH (mutant) + NADP
1ky8 - TtGAPDH + NADP – Thermoproteus tenax
3rhh - BhGAPDH + NADP
NADP-dependent GAPDH ternary complexes containing glyceraldehyde-3-phosphate (G3P)NADP-dependent GAPDH ternary complexes containing glyceraldehyde-3-phosphate (G3P)
1qi1 - SmGAPDH + NADP + G3P
2esd, 2qe0 - SmGAPDH (mutant) + NADP + G3P
3lc2 – SaGAPDH-thioacyl + G3P
1uxu - TtGAPDH (mutant) + NADP + AMP + G3P
Other NADP-dependent GAPDH ternary complexesOther NADP-dependent GAPDH ternary complexes
1k3t - TcGAPDH + coumarin derivative – Trypanosoma cruzi
1uxv - TtGAPDH + NADP + AMP
1uxp - TtGAPDH (mutant) + NADP + ADP
1uxq - TtGAPDH (mutant) + NADP + G1P
1uxr - TtGAPDH (mutant) + NADP + F6P
ReferencesReferences
1) Voet, D, Voet, J, & Pratt, C. (2008). Fundamentals of biochemistry, third edition. Hoboken, NJ: Wiley & Sons, Inc.
2)Family: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain. Retrieved from: http://scop.mrc-lmb.cam.ac.uk/scop/data/scop.b.d.c.b.d.html
3) [xtra 1]
4) [xtra 1]
- ↑ Vospelnikova ND, Safronova MI, Shuvalova ER, Baratova LA, Kniazev SP, Nagradova NK. Identification of an arginine residue important for catalytic activity in the primary structure of D-glyceraldehyde 3-phosphate dehydrogenase. Studies with the rat skeletal-muscle enzyme. Biochem J. 1981 Dec 1;199(3):757-65. PMID:7340828
5) [xtra 1]
- ↑ Butterfield DA, Hardas SS, Lange ML. Oxidatively modified glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Alzheimer's disease: many pathways to neurodegeneration. J Alzheimers Dis. 2010;20(2):369-93. PMID:20164570 doi:10.3233/JAD-2010-1375
6) [xtra 1]
- ↑ Seidler NW. Dynamic oligomeric properties. Adv Exp Med Biol. 2013;985:207-47. doi: 10.1007/978-94-007-4716-6_7. PMID:22851451 doi:http://dx.doi.org/10.1007/978-94-007-4716-6_7