Photosystem II: Difference between revisions
Michal Harel (talk | contribs) No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
<StructureSection load='3a0b' size='400' caption='Photosystem II, [[3a0b]]' scene='' > | <StructureSection load='3a0b' size='400' caption='Photosystem II, [[3a0b]]' scene='' > | ||
==Background== | ==Background== | ||
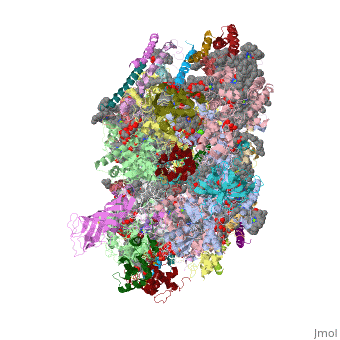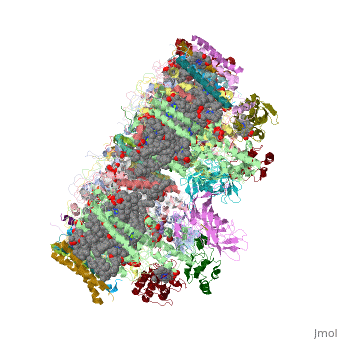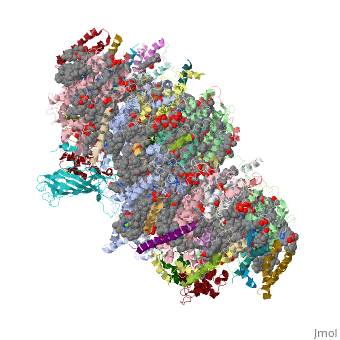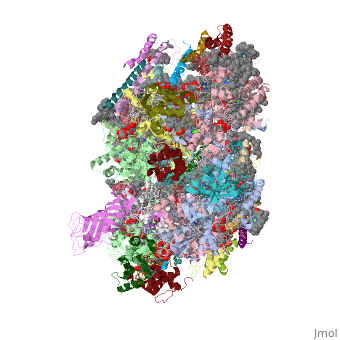
This structure of '''Photosystem II''' was crystallized from the cyanobacteria, ''Thermosynechococcus elongatus'', at 3.0Å <ref>PMID: 16355230</ref> and at 3.50 Å <ref>PMID: 14764885</ref>. PDB codes are [[2axt]] and [[1s5l]], respectively. Cyanobacteria and plants both contain Photosystem II while photosynthetic bacteria contain the bacterial reaction center. This photosynthetic protein complex is associated with a variety of functional ligands. It is a <scene name='Photosystem_II/Psii_dimer/1'>dimer</scene> composed mainly of alpha-helices. Nineteen <scene name='Photosystem_II/Protein_only/1'>subunits</scene> are in each monomer, with multiple extrinsic subunits associated with the oxygen evolving complex missing from this crystallization. Photosystem II is a membrane bound protein complex that in plants is associated with the thylakoid membrane of chloroplasts. <scene name='Photosystem_II/Hydrophobic_polar/1'>Polar and hydrophobic</scene> regions correlate with membrane associated nature of the protein. '''<FONT COLOR="#616D7E">Hydrophobic</FONT>''' helices make up the transmembranal portion, while '''<FONT COLOR="#C031C7">polar</FONT>''' residues are concentrated externally on either side of the membrane. | This structure of '''Photosystem II''' was crystallized from the cyanobacteria, ''Thermosynechococcus elongatus'', at 3.0Å <ref>PMID: 16355230</ref> and at 3.50 Å <ref>PMID: 14764885</ref>. PDB codes are [[2axt]] and [[1s5l]], respectively. Cyanobacteria and plants both contain Photosystem II while photosynthetic bacteria contain the bacterial reaction center. This photosynthetic protein complex is associated with a variety of functional ligands. It is a <scene name='Photosystem_II/Psii_dimer/1'>dimer</scene> composed mainly of alpha-helices. Nineteen <scene name='Photosystem_II/Protein_only/1'>subunits</scene> are in each monomer, with multiple extrinsic subunits associated with the oxygen evolving complex missing from this crystallization. Photosystem II is a membrane bound protein complex that in plants is associated with the thylakoid membrane of chloroplasts. <scene name='Photosystem_II/Hydrophobic_polar/1'>Polar and hydrophobic</scene> regions correlate with membrane associated nature of the protein. '''<FONT COLOR="#616D7E">Hydrophobic</FONT>''' helices make up the transmembranal portion, while '''<FONT COLOR="#C031C7">polar</FONT>''' residues are concentrated externally on either side of the membrane. | ||
Revision as of 09:45, 20 August 2014
BackgroundThis structure of Photosystem II was crystallized from the cyanobacteria, Thermosynechococcus elongatus, at 3.0Å [1] and at 3.50 Å [2]. PDB codes are 2axt and 1s5l, respectively. Cyanobacteria and plants both contain Photosystem II while photosynthetic bacteria contain the bacterial reaction center. This photosynthetic protein complex is associated with a variety of functional ligands. It is a composed mainly of alpha-helices. Nineteen are in each monomer, with multiple extrinsic subunits associated with the oxygen evolving complex missing from this crystallization. Photosystem II is a membrane bound protein complex that in plants is associated with the thylakoid membrane of chloroplasts. regions correlate with membrane associated nature of the protein. Hydrophobic helices make up the transmembranal portion, while polar residues are concentrated externally on either side of the membrane. PhotosynthesisPhotosystem II is an integral part of photosynthesis, the conversion of light energy into chemical energy by living organisms. Photosystem II is linked to a variety of other proteins, including Photosytem I. These proteins ultimately produce NADPH and ATP that power the Calvin cycle. Using this energy, glucose is synthesized from carbon dioxide and water. Electron Transfer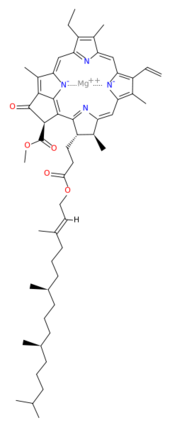 surround Photosystem II and capture energy from sunlight, exciting electrons. Chlorophyll are highly conjugated and absorb visible light, along with accessory light harvesting pigments such as . Beta carotene absorbs visible light of other wavelengths and also protects Photosystem II by destroying reactive oxygen species that result from this photoexcitation. Electrons are passed from chlorophyll to . Pheophytin are very similar to chlorophyll except they contain 2 H+ instead of a Mg2+ ion. From the pheophytin, electrons transferred to , which are reduced. Located between each pair of quinones, an iron helps to transfer the electron. These plastoquinones eventually move to a plastoquinone pool which travels to another large protein subunit, cytochrome b 6/ f. Eventually these electrons reduce NADP+ to NADPH. The through Photosystem II is shown, with beta-carotenes, pheophytins, iron and plasotoquinones. Oxygen EvolutionAnother important facet of Photosystem II is its ability to oxidize water to oxygen with its . These centers are structures with 3 manganese, 4 oxygen and a calcium linked to a fourth manganese[3]. Oxidation of water leaves 2 H + on the lumenal side of the membrane, helping to establish the proton gradient essential for ATP synthesis in the CF1CF0-ATP sythase protein. 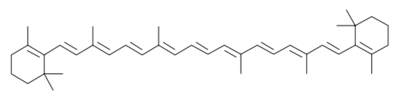 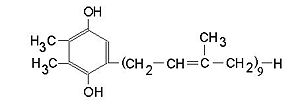 |
| ||||||||||
3D structures of photosystem II3D structures of photosystem II
Updated on 20-August-2014
3arc, 3a0b, 3a0h, 4il6 – PSII – Thermosynechococcus vulcanos
3prq, 3prr - TePSII + terbutryn – Thermosynechococcus elongatus
3kzi, 3bz1, 3bz2, 2axt, 1w5c, 1s5l, 1izl, 1ilx, 1fe1, 4fby, 4ixr, 4ixq – TePSII
3zpn - TePSII PSB28 protein
4k7b - PSII extrinsic protein – Chaetoceros gracilis
2y6x – TePSII PSB27 protein
2kvo – SyPSII reaction center PSB28 protein – Synechocystis – NMR
2kmf, 2knd - SyPSII reaction center PSB27 subunit – NMR
2vu4, 1vyk – spPSII PSBP subunit – spinach
1nze - spPSII PSBQ subunit
1v2b - PSII PSBP subunit – tobacco
1fc6, 1fc7, 1fc9, 1fcf – PSII C terminal processing protease – Scenedesmus obliquus
Additional ResourcesAdditional Resources
For additional information, see: Photosynthesis
ReferencesReferences
- ↑ Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Towards complete cofactor arrangement in the 3.0 A resolution structure of photosystem II. Nature. 2005 Dec 15;438(7070):1040-4. PMID:16355230 doi:http://dx.doi.org/10.1038/nature04224
- ↑ Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004 Mar 19;303(5665):1831-8. Epub 2004 Feb 5. PMID:14764885 doi:http://dx.doi.org/10.1126/science.1093087
- ↑ Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Architecture of the photosynthetic oxygen-evolving center. Science. 2004 Mar 19;303(5665):1831-8. Epub 2004 Feb 5. PMID:14764885 doi:http://dx.doi.org/10.1126/science.1093087