1ihr: Difference between revisions
No edit summary |
No edit summary |
||
| (9 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
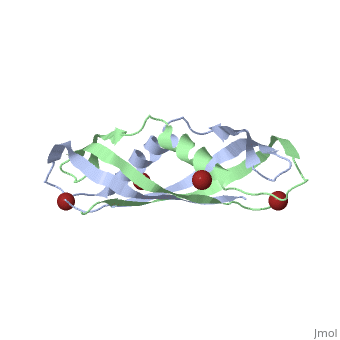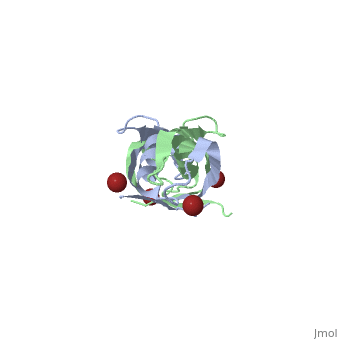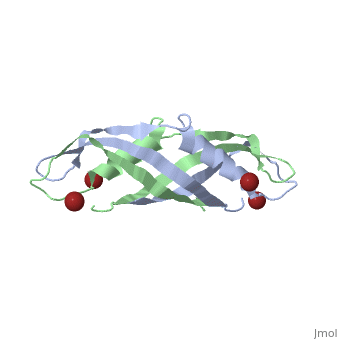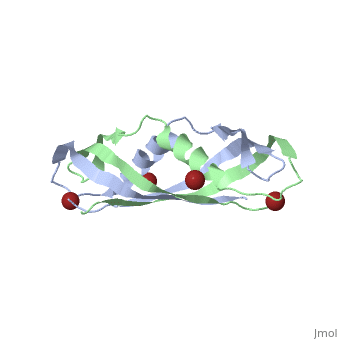
< | ==Crystal structure of the dimeric C-terminal domain of TonB== | ||
<StructureSection load='1ihr' size='340' side='right'caption='[[1ihr]], [[Resolution|resolution]] 1.55Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1ihr]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Escherichia_coli Escherichia coli]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1IHR OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1IHR FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 1.55Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=BR:BROMIDE+ION'>BR</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1ihr FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1ihr OCA], [https://pdbe.org/1ihr PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1ihr RCSB], [https://www.ebi.ac.uk/pdbsum/1ihr PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1ihr ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/TONB_ECOLI TONB_ECOLI] Interacts with outer membrane receptor proteins that carry out high-affinity binding and energy dependent uptake into the periplasmic space of specific substrates such as cobalamin, and various iron compounds (such as iron dicitrate, enterochelin, aerobactin, etc.). In the absence of TonB these receptors bind their substrates but do not carry out active transport. TonB also interacts with some colicins and is involved in the energy-dependent, irreversible steps of bacteriophages phi 80 and T1 infection. It could act to transduce energy from the cytoplasmic membrane to specific energy-requiring processes in the outer membrane, resulting in the release into the periplasm of ligands bound by these outer membrane proteins. | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/ih/1ihr_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1ihr ConSurf]. | |||
<div style="clear:both"></div> | |||
== | ==See Also== | ||
*[[TonB|TonB]] | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Escherichia coli]] | [[Category: Escherichia coli]] | ||
[[Category: | [[Category: Large Structures]] | ||
[[Category: | [[Category: Chang C]] | ||
[[Category: | [[Category: Mooser A]] | ||
[[Category: | [[Category: Pluckthun A]] | ||
[[Category: | [[Category: Wlodawer A]] | ||
Latest revision as of 10:35, 7 February 2024
Crystal structure of the dimeric C-terminal domain of TonBCrystal structure of the dimeric C-terminal domain of TonB
Structural highlights
FunctionTONB_ECOLI Interacts with outer membrane receptor proteins that carry out high-affinity binding and energy dependent uptake into the periplasmic space of specific substrates such as cobalamin, and various iron compounds (such as iron dicitrate, enterochelin, aerobactin, etc.). In the absence of TonB these receptors bind their substrates but do not carry out active transport. TonB also interacts with some colicins and is involved in the energy-dependent, irreversible steps of bacteriophages phi 80 and T1 infection. It could act to transduce energy from the cytoplasmic membrane to specific energy-requiring processes in the outer membrane, resulting in the release into the periplasm of ligands bound by these outer membrane proteins. Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. See Also |
| ||||||||||||||||||