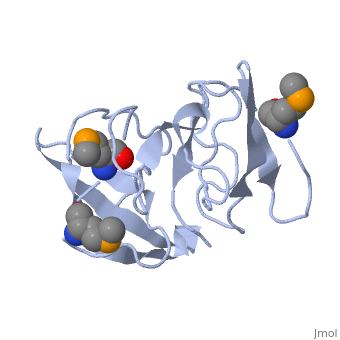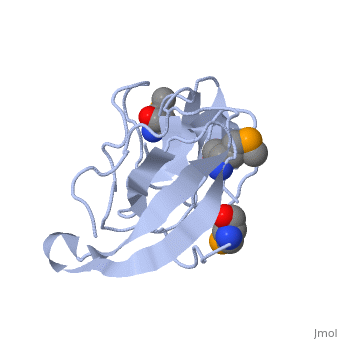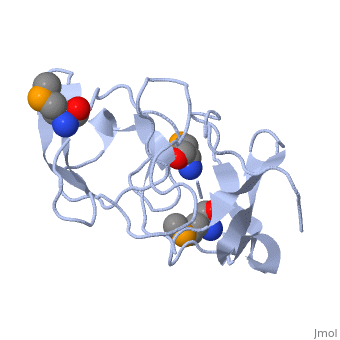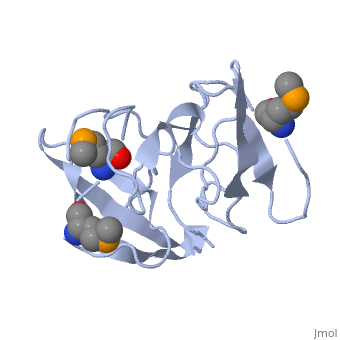1rl2: Difference between revisions
No edit summary |
No edit summary |
||
| (12 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
< | ==RIBOSOMAL PROTEIN L2 RNA-BINDING DOMAIN FROM BACILLUS STEAROTHERMOPHILUS== | ||
<StructureSection load='1rl2' size='340' side='right'caption='[[1rl2]], [[Resolution|resolution]] 2.30Å' scene=''> | |||
You may | == Structural highlights == | ||
or the | <table><tr><td colspan='2'>[[1rl2]] is a 2 chain structure with sequence from [https://en.wikipedia.org/wiki/Geobacillus_stearothermophilus Geobacillus stearothermophilus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1RL2 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1RL2 FirstGlance]. <br> | ||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.3Å</td></tr> | |||
-- | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=MSE:SELENOMETHIONINE'>MSE</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1rl2 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1rl2 OCA], [https://pdbe.org/1rl2 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1rl2 RCSB], [https://www.ebi.ac.uk/pdbsum/1rl2 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1rl2 ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/RL2_GEOSE RL2_GEOSE] One of the primary rRNA binding proteins. Required for association of the 30S and 50S subunits to form the 70S ribosome, for tRNA binding and peptide bond formation. It has been suggested to have peptidyltransferase activity; this is somewhat controversial. Makes several contacts with the 16S rRNA in the 70S ribosome (By similarity).[HAMAP-Rule:MF_01320] | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/rl/1rl2_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1rl2 ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Ribosomal protein L2 is the largest protein component in the ribosome. It is located at or near the peptidyl transferase center and has been a prime candidate for the peptidyl transferase activity. It binds directly to 23S rRNA and plays a crucial role in its assembly. The three-dimensional structure of the RNA-binding domain of L2 from Bacillus stearothermophilus has been determined at 2.3 A resolution by X-ray crystallography using the selenomethionyl MAD method. The RNA-binding domain of L2 consists of two recurring motifs of approximately 70 residues each. The N-terminal domain (positions 60-130) is homologous to the OB-fold, and the C-terminal domain (positions 131-201) is homologous to the SH3-like barrel. Residues Arg86 and Arg155, which have been identified by mutation experiments to be involved in the 23S rRNA binding, are located at the gate of the interface region between the two domains. The molecular architecture suggests how this important protein has evolved from the ancient nucleic acid-binding proteins to create a 23S rRNA-binding domain in the very remote past. | |||
The three-dimensional structure of the RNA-binding domain of ribosomal protein L2; a protein at the peptidyl transferase center of the ribosome.,Nakagawa A, Nakashima T, Taniguchi M, Hosaka H, Kimura M, Tanaka I EMBO J. 1999 Mar 15;18(6):1459-67. PMID:10075918<ref>PMID:10075918</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 1rl2" style="background-color:#fffaf0;"></div> | |||
==See Also== | |||
*[[Ribosomal protein L2|Ribosomal protein L2]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
== | |||
== | |||
[[Category: Geobacillus stearothermophilus]] | [[Category: Geobacillus stearothermophilus]] | ||
[[Category: | [[Category: Large Structures]] | ||
[[Category: Hosaka | [[Category: Hosaka H]] | ||
[[Category: Nakagawa | [[Category: Nakagawa A]] | ||
[[Category: Nakashima | [[Category: Nakashima T]] | ||
[[Category: Tanaka | [[Category: Tanaka I]] | ||
Latest revision as of 03:27, 21 November 2024
RIBOSOMAL PROTEIN L2 RNA-BINDING DOMAIN FROM BACILLUS STEAROTHERMOPHILUSRIBOSOMAL PROTEIN L2 RNA-BINDING DOMAIN FROM BACILLUS STEAROTHERMOPHILUS
Structural highlights
FunctionRL2_GEOSE One of the primary rRNA binding proteins. Required for association of the 30S and 50S subunits to form the 70S ribosome, for tRNA binding and peptide bond formation. It has been suggested to have peptidyltransferase activity; this is somewhat controversial. Makes several contacts with the 16S rRNA in the 70S ribosome (By similarity).[HAMAP-Rule:MF_01320] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedRibosomal protein L2 is the largest protein component in the ribosome. It is located at or near the peptidyl transferase center and has been a prime candidate for the peptidyl transferase activity. It binds directly to 23S rRNA and plays a crucial role in its assembly. The three-dimensional structure of the RNA-binding domain of L2 from Bacillus stearothermophilus has been determined at 2.3 A resolution by X-ray crystallography using the selenomethionyl MAD method. The RNA-binding domain of L2 consists of two recurring motifs of approximately 70 residues each. The N-terminal domain (positions 60-130) is homologous to the OB-fold, and the C-terminal domain (positions 131-201) is homologous to the SH3-like barrel. Residues Arg86 and Arg155, which have been identified by mutation experiments to be involved in the 23S rRNA binding, are located at the gate of the interface region between the two domains. The molecular architecture suggests how this important protein has evolved from the ancient nucleic acid-binding proteins to create a 23S rRNA-binding domain in the very remote past. The three-dimensional structure of the RNA-binding domain of ribosomal protein L2; a protein at the peptidyl transferase center of the ribosome.,Nakagawa A, Nakashima T, Taniguchi M, Hosaka H, Kimura M, Tanaka I EMBO J. 1999 Mar 15;18(6):1459-67. PMID:10075918[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||