1gzr: Difference between revisions
No edit summary |
No edit summary |
||
| (15 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Human Insulin-like growth factor; ESRF data== | |||
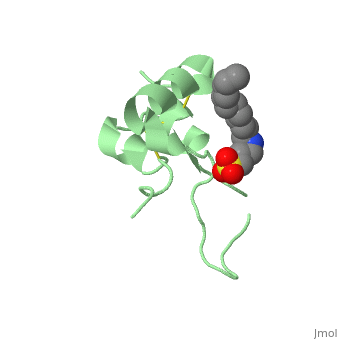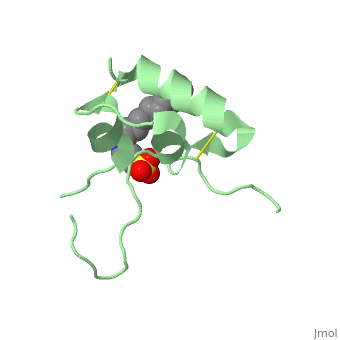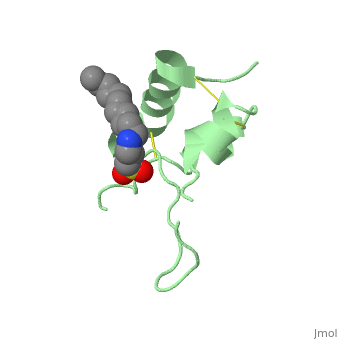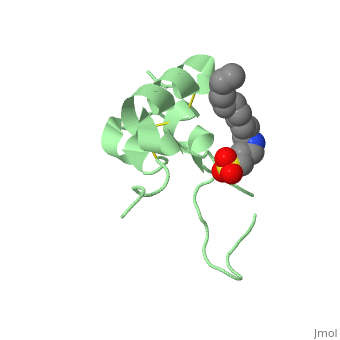
<StructureSection load='1gzr' size='340' side='right'caption='[[1gzr]], [[Resolution|resolution]] 2.00Å' scene=''> | |||
== Structural highlights == | |||
| | <table><tr><td colspan='2'>[[1gzr]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1GZR OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1GZR FirstGlance]. <br> | ||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=C15:N-DODECYL-N,N-DIMETHYL-3-AMMONIO-1-PROPANESULFONATE'>C15</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1gzr FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1gzr OCA], [https://pdbe.org/1gzr PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1gzr RCSB], [https://www.ebi.ac.uk/pdbsum/1gzr PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1gzr ProSAT]</span></td></tr> | |||
</table> | |||
== Disease == | |||
[https://www.uniprot.org/uniprot/IGF1_HUMAN IGF1_HUMAN] Defects in IGF1 are the cause of insulin-like growth factor I deficiency (IGF1 deficiency) [MIM:[https://omim.org/entry/608747 608747]. IGF1 deficiency is an autosomal recessive disorder characterized by growth retardation, sensorineural deafness and mental retardation. | |||
== Function == | |||
== | [https://www.uniprot.org/uniprot/IGF1_HUMAN IGF1_HUMAN] The insulin-like growth factors, isolated from plasma, are structurally and functionally related to insulin but have a much higher growth-promoting activity. May be a physiological regulator of [1-14C]-2-deoxy-D-glucose (2DG) transport and glycogen synthesis in osteoblasts. Stimulates glucose transport in rat bone-derived osteoblastic (PyMS) cells and is effective at much lower concentrations than insulin, not only regarding glycogen and DNA synthesis but also with regard to enhancing glucose uptake.<ref>PMID:21076856</ref> | ||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/gz/1gzr_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1gzr ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Human insulin-like growth factors I and II (hIGF-I, hIGF-II) are potent stimulators of cell and growth processes. They display high sequence similarity to both the A and B chains of insulin but contain an additional connecting C-domain, which reflects their secretion without specific packaging or precursor conversion. IGFs also have an extension at the C-terminus known as the D-domain. This paper describes four homologous hIGF-1 structures, obtained from crystals grown in the presence of the detergent SB12, which reveal additional detail in the C- and D-domains. Two different detergent binding modes observed in the crystals may reflect different hIGF-I biological properties such as the interaction with IGF binding proteins and self-aggregation. While the helical core of hIGF-I is very similar to that in insulin, there are distinct differences in the region of hIGF-I corresponding to the insulin B chain C-terminus, residues B25-B30. In hIGF-I, these residues (24-29) and the following C-domain form an extensive loop protruding 20 A from the core, which results in a substantially different conformation for the receptor binding epitope in hIGF-I compared to insulin. One notable feature of the structures presented here is demonstration of peptide-bond cleavage between Ser35 and Arg36 resulting in an apparent gap between residues 35 and 39. The equivalent region of proinsulin is involved in hormone processing demanding a reassessment of the structural integrity of hIGF-I in relation to its biological function. | Human insulin-like growth factors I and II (hIGF-I, hIGF-II) are potent stimulators of cell and growth processes. They display high sequence similarity to both the A and B chains of insulin but contain an additional connecting C-domain, which reflects their secretion without specific packaging or precursor conversion. IGFs also have an extension at the C-terminus known as the D-domain. This paper describes four homologous hIGF-1 structures, obtained from crystals grown in the presence of the detergent SB12, which reveal additional detail in the C- and D-domains. Two different detergent binding modes observed in the crystals may reflect different hIGF-I biological properties such as the interaction with IGF binding proteins and self-aggregation. While the helical core of hIGF-I is very similar to that in insulin, there are distinct differences in the region of hIGF-I corresponding to the insulin B chain C-terminus, residues B25-B30. In hIGF-I, these residues (24-29) and the following C-domain form an extensive loop protruding 20 A from the core, which results in a substantially different conformation for the receptor binding epitope in hIGF-I compared to insulin. One notable feature of the structures presented here is demonstration of peptide-bond cleavage between Ser35 and Arg36 resulting in an apparent gap between residues 35 and 39. The equivalent region of proinsulin is involved in hormone processing demanding a reassessment of the structural integrity of hIGF-I in relation to its biological function. | ||
Structural origins of the functional divergence of human insulin-like growth factor-I and insulin.,Brzozowski AM, Dodson EJ, Dodson GG, Murshudov GN, Verma C, Turkenburg JP, de Bree FM, Dauter Z Biochemistry. 2002 Jul 30;41(30):9389-97. PMID:12135360<ref>PMID:12135360</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 1gzr" style="background-color:#fffaf0;"></div> | |||
== | ==See Also== | ||
*[[Insulin-like growth factor|Insulin-like growth factor]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: | [[Category: Large Structures]] | ||
[[Category: | [[Category: Brzozowski AM]] | ||
[[Category: | [[Category: Dauter Z]] | ||
[[Category: | [[Category: De Bree FM]] | ||
[[Category: Dodson | [[Category: Dodson EJ]] | ||
[[Category: Dodson | [[Category: Dodson GG]] | ||
[[Category: Murshudov | [[Category: Murshudov G]] | ||
[[Category: Turkenburg | [[Category: Turkenburg JP]] | ||
[[Category: Verma | [[Category: Verma C]] | ||
Latest revision as of 11:29, 6 November 2024
Human Insulin-like growth factor; ESRF dataHuman Insulin-like growth factor; ESRF data
Structural highlights
DiseaseIGF1_HUMAN Defects in IGF1 are the cause of insulin-like growth factor I deficiency (IGF1 deficiency) [MIM:608747. IGF1 deficiency is an autosomal recessive disorder characterized by growth retardation, sensorineural deafness and mental retardation. FunctionIGF1_HUMAN The insulin-like growth factors, isolated from plasma, are structurally and functionally related to insulin but have a much higher growth-promoting activity. May be a physiological regulator of [1-14C]-2-deoxy-D-glucose (2DG) transport and glycogen synthesis in osteoblasts. Stimulates glucose transport in rat bone-derived osteoblastic (PyMS) cells and is effective at much lower concentrations than insulin, not only regarding glycogen and DNA synthesis but also with regard to enhancing glucose uptake.[1] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedHuman insulin-like growth factors I and II (hIGF-I, hIGF-II) are potent stimulators of cell and growth processes. They display high sequence similarity to both the A and B chains of insulin but contain an additional connecting C-domain, which reflects their secretion without specific packaging or precursor conversion. IGFs also have an extension at the C-terminus known as the D-domain. This paper describes four homologous hIGF-1 structures, obtained from crystals grown in the presence of the detergent SB12, which reveal additional detail in the C- and D-domains. Two different detergent binding modes observed in the crystals may reflect different hIGF-I biological properties such as the interaction with IGF binding proteins and self-aggregation. While the helical core of hIGF-I is very similar to that in insulin, there are distinct differences in the region of hIGF-I corresponding to the insulin B chain C-terminus, residues B25-B30. In hIGF-I, these residues (24-29) and the following C-domain form an extensive loop protruding 20 A from the core, which results in a substantially different conformation for the receptor binding epitope in hIGF-I compared to insulin. One notable feature of the structures presented here is demonstration of peptide-bond cleavage between Ser35 and Arg36 resulting in an apparent gap between residues 35 and 39. The equivalent region of proinsulin is involved in hormone processing demanding a reassessment of the structural integrity of hIGF-I in relation to its biological function. Structural origins of the functional divergence of human insulin-like growth factor-I and insulin.,Brzozowski AM, Dodson EJ, Dodson GG, Murshudov GN, Verma C, Turkenburg JP, de Bree FM, Dauter Z Biochemistry. 2002 Jul 30;41(30):9389-97. PMID:12135360[2] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||