1l6j: Difference between revisions
No edit summary |
No edit summary |
||
| (18 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
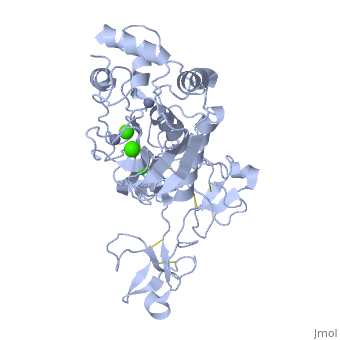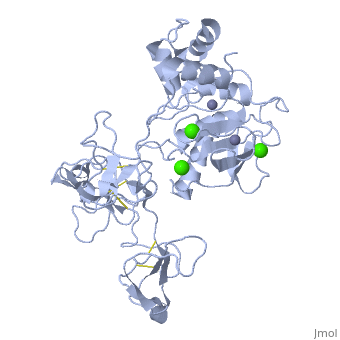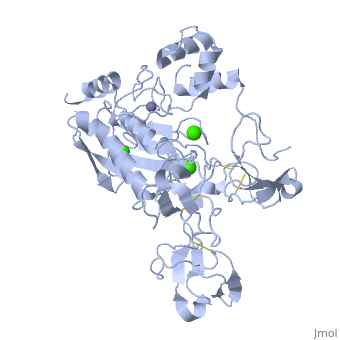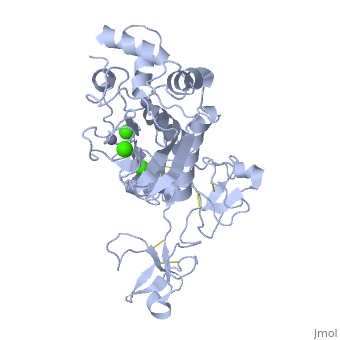
== | ==Crystal structure of human matrix metalloproteinase MMP9 (gelatinase B).== | ||
<StructureSection load='1l6j' size='340' side='right'caption='[[1l6j]], [[Resolution|resolution]] 2.50Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1l6j]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1L6J OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1L6J FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.5Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CA:CALCIUM+ION'>CA</scene>, <scene name='pdbligand=ZN:ZINC+ION'>ZN</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1l6j FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1l6j OCA], [https://pdbe.org/1l6j PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1l6j RCSB], [https://www.ebi.ac.uk/pdbsum/1l6j PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1l6j ProSAT]</span></td></tr> | |||
</table> | |||
== Disease == | |||
[https://www.uniprot.org/uniprot/MMP9_HUMAN MMP9_HUMAN] Defects in MMP9 may be a cause of susceptibility to intervertebral disc disease (IDD) [MIM:[https://omim.org/entry/603932 603932]; also known as lumbar disk herniation (LDH). IDD is one of the most common musculo-skeletal disorders and the predominant cause of low-back pain and unilateral leg pain.<ref>PMID:18455130</ref> Defects in MMP9 are the cause of metaphyseal anadysplasia type 2 (MANDP2) [MIM:[https://omim.org/entry/613073 613073]. Metaphyseal anadysplasia consists of an abnormal bone development characterized by severe skeletal changes that, in contrast with the progressive course of most other skeletal dysplasias, resolve spontaneously with age. Clinical characteristics are evident from the first months of life and include slight shortness of stature and a mild varus deformity of the legs. Patients attain a normal stature in adolescence and show improvement or complete resolution of varus deformity of the legs and rhizomelic micromelia. | |||
== Function == | |||
[https://www.uniprot.org/uniprot/MMP9_HUMAN MMP9_HUMAN] May play an essential role in local proteolysis of the extracellular matrix and in leukocyte migration. Could play a role in bone osteoclastic resorption. Cleaves KiSS1 at a Gly-|-Leu bond. Cleaves type IV and type V collagen into large C-terminal three quarter fragments and shorter N-terminal one quarter fragments. Degrades fibronectin but not laminin or Pz-peptide.<ref>PMID:1480034</ref> | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/l6/1l6j_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview03.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1l6j ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
The X-ray crystal structure of the proform of human matrix metalloproteinase MMP9 has been solved to 2.5 A resolution. The construct includes the prodomain, the catalytic domain and three FnII (fibronectin type II) domains. The prodomain is inserted into the active-site cleft, blocking access to the catalytic zinc. Comparison with the crystal structure of the most closely related MMP, MMP2, indicates that the conformations of residues in the active-site cleft and in the cysteine-switch peptide of the prodomain are highly conserved and that design of MMP9-specific inhibitors will be challenging. In common with MMP2, the MMP9 S1' inhibitor-binding pocket is large compared with that of other MMPs. One small point of difference in the S1' binding pockets of MMP9 and MMP2 may provide an opportunity to explore the design of specific inhibitors. The side chain of Arg424 in MMP9 is angled slightly away from the S1' pocket when compared with the corresponding residue in MMP2, Thr424. The secondary structure of the FnII domains is conserved between the two closely related MMPs, although the second FnII domain makes no contact with the catalytic domain in MMP9, while the same domain in MMP2 has a substantial area of interaction with the catalytic domain. | The X-ray crystal structure of the proform of human matrix metalloproteinase MMP9 has been solved to 2.5 A resolution. The construct includes the prodomain, the catalytic domain and three FnII (fibronectin type II) domains. The prodomain is inserted into the active-site cleft, blocking access to the catalytic zinc. Comparison with the crystal structure of the most closely related MMP, MMP2, indicates that the conformations of residues in the active-site cleft and in the cysteine-switch peptide of the prodomain are highly conserved and that design of MMP9-specific inhibitors will be challenging. In common with MMP2, the MMP9 S1' inhibitor-binding pocket is large compared with that of other MMPs. One small point of difference in the S1' binding pockets of MMP9 and MMP2 may provide an opportunity to explore the design of specific inhibitors. The side chain of Arg424 in MMP9 is angled slightly away from the S1' pocket when compared with the corresponding residue in MMP2, Thr424. The secondary structure of the FnII domains is conserved between the two closely related MMPs, although the second FnII domain makes no contact with the catalytic domain in MMP9, while the same domain in MMP2 has a substantial area of interaction with the catalytic domain. | ||
Structure of the C-terminally truncated human ProMMP9, a gelatin-binding matrix metalloproteinase.,Elkins PA, Ho YS, Smith WW, Janson CA, D'Alessio KJ, McQueney MS, Cummings MD, Romanic AM Acta Crystallogr D Biol Crystallogr. 2002 Jul;58(Pt 7):1182-92. Epub 2002, Jun 20. PMID:12077439<ref>PMID:12077439</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
[[ | <div class="pdbe-citations 1l6j" style="background-color:#fffaf0;"></div> | ||
==See Also== | |||
*[[Matrix metalloproteinase 3D structures|Matrix metalloproteinase 3D structures]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Homo sapiens]] | [[Category: Homo sapiens]] | ||
[[Category: | [[Category: Large Structures]] | ||
[[Category: | [[Category: Cummings MD]] | ||
[[Category: | [[Category: D'Alessio KJ]] | ||
[[Category: Elkins | [[Category: Elkins PA]] | ||
[[Category: Ho | [[Category: Ho YS]] | ||
[[Category: Janson | [[Category: Janson CA]] | ||
[[Category: McQueney | [[Category: McQueney MS]] | ||
[[Category: Romanic | [[Category: Romanic AM]] | ||
[[Category: Smith | [[Category: Smith WW]] | ||
Latest revision as of 07:41, 17 October 2024
Crystal structure of human matrix metalloproteinase MMP9 (gelatinase B).Crystal structure of human matrix metalloproteinase MMP9 (gelatinase B).
Structural highlights
DiseaseMMP9_HUMAN Defects in MMP9 may be a cause of susceptibility to intervertebral disc disease (IDD) [MIM:603932; also known as lumbar disk herniation (LDH). IDD is one of the most common musculo-skeletal disorders and the predominant cause of low-back pain and unilateral leg pain.[1] Defects in MMP9 are the cause of metaphyseal anadysplasia type 2 (MANDP2) [MIM:613073. Metaphyseal anadysplasia consists of an abnormal bone development characterized by severe skeletal changes that, in contrast with the progressive course of most other skeletal dysplasias, resolve spontaneously with age. Clinical characteristics are evident from the first months of life and include slight shortness of stature and a mild varus deformity of the legs. Patients attain a normal stature in adolescence and show improvement or complete resolution of varus deformity of the legs and rhizomelic micromelia. FunctionMMP9_HUMAN May play an essential role in local proteolysis of the extracellular matrix and in leukocyte migration. Could play a role in bone osteoclastic resorption. Cleaves KiSS1 at a Gly-|-Leu bond. Cleaves type IV and type V collagen into large C-terminal three quarter fragments and shorter N-terminal one quarter fragments. Degrades fibronectin but not laminin or Pz-peptide.[2] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe X-ray crystal structure of the proform of human matrix metalloproteinase MMP9 has been solved to 2.5 A resolution. The construct includes the prodomain, the catalytic domain and three FnII (fibronectin type II) domains. The prodomain is inserted into the active-site cleft, blocking access to the catalytic zinc. Comparison with the crystal structure of the most closely related MMP, MMP2, indicates that the conformations of residues in the active-site cleft and in the cysteine-switch peptide of the prodomain are highly conserved and that design of MMP9-specific inhibitors will be challenging. In common with MMP2, the MMP9 S1' inhibitor-binding pocket is large compared with that of other MMPs. One small point of difference in the S1' binding pockets of MMP9 and MMP2 may provide an opportunity to explore the design of specific inhibitors. The side chain of Arg424 in MMP9 is angled slightly away from the S1' pocket when compared with the corresponding residue in MMP2, Thr424. The secondary structure of the FnII domains is conserved between the two closely related MMPs, although the second FnII domain makes no contact with the catalytic domain in MMP9, while the same domain in MMP2 has a substantial area of interaction with the catalytic domain. Structure of the C-terminally truncated human ProMMP9, a gelatin-binding matrix metalloproteinase.,Elkins PA, Ho YS, Smith WW, Janson CA, D'Alessio KJ, McQueney MS, Cummings MD, Romanic AM Acta Crystallogr D Biol Crystallogr. 2002 Jul;58(Pt 7):1182-92. Epub 2002, Jun 20. PMID:12077439[3] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||