1uyn: Difference between revisions
No edit summary |
No edit summary |
||
| (8 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
==Translocator domain of autotransporter NalP from Neisseria meningitidis== | |||
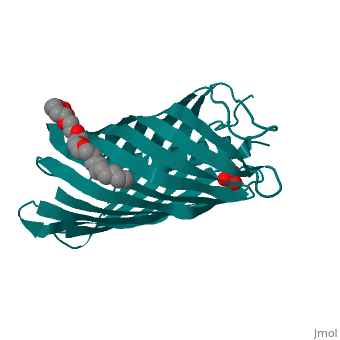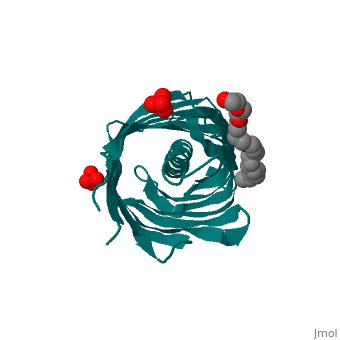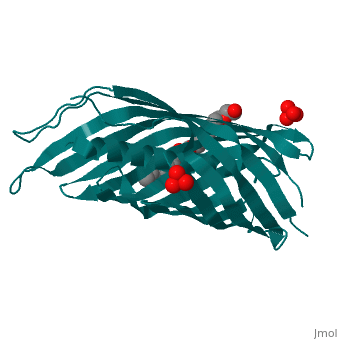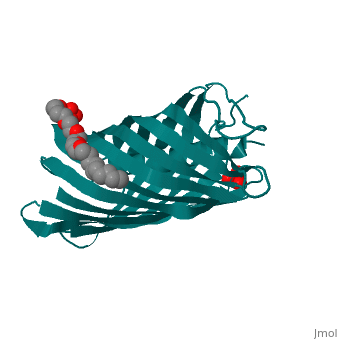
<StructureSection load='1uyn' size='340' side='right'caption='[[1uyn]], [[Resolution|resolution]] 2.60Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[1uyn]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Neisseria_meningitidis Neisseria meningitidis]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=1UYN OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=1UYN FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.6Å</td></tr> | |||
<tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CXE:PENTAETHYLENE+GLYCOL+MONODECYL+ETHER'>CXE</scene>, <scene name='pdbligand=SO4:SULFATE+ION'>SO4</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=1uyn FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=1uyn OCA], [https://pdbe.org/1uyn PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=1uyn RCSB], [https://www.ebi.ac.uk/pdbsum/1uyn PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=1uyn ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/NALP_NEIMH NALP_NEIMH] Major human immunogenic protein. Autotransporter with a secreted protease domain involved in processing other autotransporter proteins including App and IgA. Probably autoprocesses to release the about 70 kDa passenger domain (PubMed:14617158). Processes the lactoferrin receptor lipoprotein subunit (LbpB) extracellularly, releasing it from the cell surface. LbpB release protects bacteria against complement-mediated killing by anti-LbpB antibodies (PubMed:20421383). Processes NHBA (PubMed:23258267). Lipidation slows its auto-processing, probably allowing it to act on endogenous substrates on the cell suface before the passenger domain is released into the medium (PubMed:23258267). The C-terminal beta-barrel domain inserts into the outer membrane where it probably exports the N-terminal passenger domain (PubMed:15014442). Both the cell surface protein (Neisserial autotransporter lipoprotein NalP) and the passenger domain cleave human (host) complement factor C3, generating a shorter alpha chain and a longer beta chain than normal (By similarity).[UniProtKB:Q9JXM7]<ref>PMID:14617158</ref> <ref>PMID:15014442</ref> <ref>PMID:20421383</ref> <ref>PMID:23258267</ref> Plays a role in extracellular-DNA (eDNA) mediated biofilm formation. In some strains (including cc32 strain H44/76 but not cc11 strain B16B6) eDNA stimulates biofilm formation. When NalP is not expressed (and no longer processes NHBA or IgA) biofilm formation increases (PubMed:23163582). This is probably because the number of positively charged, DNA-binding peptides on the cell surface rises, resulting in increased biofilm formation (Probable).<ref>PMID:23163582</ref> <ref>PMID:23163582</ref> Cleaves human (host) complement factor C3, generating a shorter alpha chain and a longer beta chain than normal. Does not act on mouse or rabbit C3. Cleavage causes C3b degradation by human CFI and CFH, decreases deposition of C3b on the bacteria surface and probably facilitates complement escape.[UniProtKB:Q9JXM7] | |||
== Evolutionary Conservation == | |||
[[Image:Consurf_key_small.gif|200px|right]] | |||
Check<jmol> | |||
<jmolCheckbox> | |||
<scriptWhenChecked>; select protein; define ~consurf_to_do selected; consurf_initial_scene = true; script "/wiki/ConSurf/uy/1uyn_consurf.spt"</scriptWhenChecked> | |||
<scriptWhenUnchecked>script /wiki/extensions/Proteopedia/spt/initialview01.spt</scriptWhenUnchecked> | |||
<text>to colour the structure by Evolutionary Conservation</text> | |||
</jmolCheckbox> | |||
</jmol>, as determined by [http://consurfdb.tau.ac.il/ ConSurfDB]. You may read the [[Conservation%2C_Evolutionary|explanation]] of the method and the full data available from [http://bental.tau.ac.il/new_ConSurfDB/main_output.php?pdb_ID=1uyn ConSurf]. | |||
<div style="clear:both"></div> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Autotransporters are virulence-related proteins of Gram-negative bacteria that are secreted via an outer-membrane-based C-terminal extension, the translocator domain. This domain supposedly is sufficient for the transport of the N-terminal passenger domain across the outer membrane. We present here the crystal structure of the in vitro-folded translocator domain of the autotransporter NalP from Neisseria meningitidis, which reveals a 12-stranded beta-barrel with a hydrophilic pore of 10 x 12.5 A that is filled by an N-terminal alpha-helix. The domain has pore activity in vivo and in vitro. Our data are consistent with the model of passenger-domain transport through the hydrophilic channel within the beta-barrel, and inconsistent with a model for transport through a central channel formed by an oligomer of translocator domains. However, the dimensions of the pore imply translocation of the secreted domain in an unfolded form. An alternative model, possibly covering the transport of folded domains, is that passenger-domain transport involves the Omp85 complex, the machinery required for membrane insertion of outer-membrane proteins, on which autotransporters are dependent. | |||
Structure of the translocator domain of a bacterial autotransporter.,Oomen CJ, van Ulsen P, van Gelder P, Feijen M, Tommassen J, Gros P EMBO J. 2004 Mar 24;23(6):1257-66. Epub 2004 Mar 11. PMID:15014442<ref>PMID:15014442</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 1uyn" style="background-color:#fffaf0;"></div> | |||
== | ==See Also== | ||
[[ | *[[NalP|NalP]] | ||
== References == | |||
== | <references/> | ||
< | __TOC__ | ||
</StructureSection> | |||
[[Category: Large Structures]] | |||
[[Category: Neisseria meningitidis]] | [[Category: Neisseria meningitidis]] | ||
[[Category: Feijen | [[Category: Feijen M]] | ||
[[Category: Gros P]] | |||
[[Category: Gros | [[Category: Oomen CJ]] | ||
[[Category: Oomen | [[Category: Tommassen J]] | ||
[[Category: Tommassen | [[Category: Van Gelder P]] | ||
[[Category: | [[Category: Van Ulsen P]] | ||
[[Category: | |||
Latest revision as of 12:06, 9 May 2024
Translocator domain of autotransporter NalP from Neisseria meningitidisTranslocator domain of autotransporter NalP from Neisseria meningitidis
Structural highlights
FunctionNALP_NEIMH Major human immunogenic protein. Autotransporter with a secreted protease domain involved in processing other autotransporter proteins including App and IgA. Probably autoprocesses to release the about 70 kDa passenger domain (PubMed:14617158). Processes the lactoferrin receptor lipoprotein subunit (LbpB) extracellularly, releasing it from the cell surface. LbpB release protects bacteria against complement-mediated killing by anti-LbpB antibodies (PubMed:20421383). Processes NHBA (PubMed:23258267). Lipidation slows its auto-processing, probably allowing it to act on endogenous substrates on the cell suface before the passenger domain is released into the medium (PubMed:23258267). The C-terminal beta-barrel domain inserts into the outer membrane where it probably exports the N-terminal passenger domain (PubMed:15014442). Both the cell surface protein (Neisserial autotransporter lipoprotein NalP) and the passenger domain cleave human (host) complement factor C3, generating a shorter alpha chain and a longer beta chain than normal (By similarity).[UniProtKB:Q9JXM7][1] [2] [3] [4] Plays a role in extracellular-DNA (eDNA) mediated biofilm formation. In some strains (including cc32 strain H44/76 but not cc11 strain B16B6) eDNA stimulates biofilm formation. When NalP is not expressed (and no longer processes NHBA or IgA) biofilm formation increases (PubMed:23163582). This is probably because the number of positively charged, DNA-binding peptides on the cell surface rises, resulting in increased biofilm formation (Probable).[5] [6] Cleaves human (host) complement factor C3, generating a shorter alpha chain and a longer beta chain than normal. Does not act on mouse or rabbit C3. Cleavage causes C3b degradation by human CFI and CFH, decreases deposition of C3b on the bacteria surface and probably facilitates complement escape.[UniProtKB:Q9JXM7] Evolutionary Conservation Check, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedAutotransporters are virulence-related proteins of Gram-negative bacteria that are secreted via an outer-membrane-based C-terminal extension, the translocator domain. This domain supposedly is sufficient for the transport of the N-terminal passenger domain across the outer membrane. We present here the crystal structure of the in vitro-folded translocator domain of the autotransporter NalP from Neisseria meningitidis, which reveals a 12-stranded beta-barrel with a hydrophilic pore of 10 x 12.5 A that is filled by an N-terminal alpha-helix. The domain has pore activity in vivo and in vitro. Our data are consistent with the model of passenger-domain transport through the hydrophilic channel within the beta-barrel, and inconsistent with a model for transport through a central channel formed by an oligomer of translocator domains. However, the dimensions of the pore imply translocation of the secreted domain in an unfolded form. An alternative model, possibly covering the transport of folded domains, is that passenger-domain transport involves the Omp85 complex, the machinery required for membrane insertion of outer-membrane proteins, on which autotransporters are dependent. Structure of the translocator domain of a bacterial autotransporter.,Oomen CJ, van Ulsen P, van Gelder P, Feijen M, Tommassen J, Gros P EMBO J. 2004 Mar 24;23(6):1257-66. Epub 2004 Mar 11. PMID:15014442[7] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||