3zr6: Difference between revisions
No edit summary |
No edit summary |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
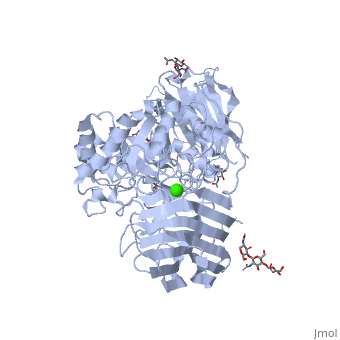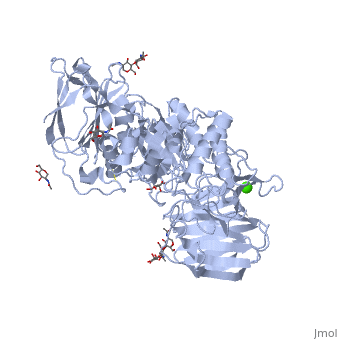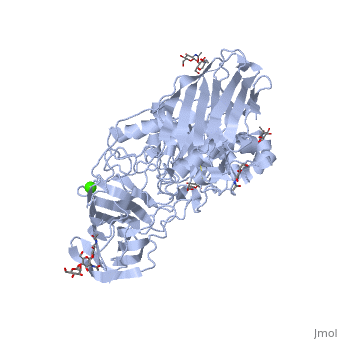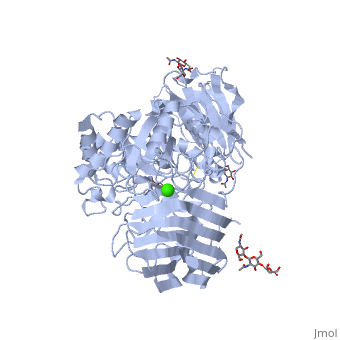
< | ==STRUCTURE OF GALACTOCEREBROSIDASE FROM MOUSE IN COMPLEX WITH GALACTOSE== | ||
<StructureSection load='3zr6' size='340' side='right'caption='[[3zr6]], [[Resolution|resolution]] 2.44Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[3zr6]] is a 1 chain structure with sequence from [https://en.wikipedia.org/wiki/Mus_musculus Mus musculus]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=3ZR6 OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=3ZR6 FirstGlance]. <br> | |||
</td></tr><tr id='method'><td class="sblockLbl"><b>[[Empirical_models|Method:]]</b></td><td class="sblockDat" id="methodDat">X-ray diffraction, [[Resolution|Resolution]] 2.44Å</td></tr> | |||
- | <tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=BMA:BETA-D-MANNOSE'>BMA</scene>, <scene name='pdbligand=CA:CALCIUM+ION'>CA</scene>, <scene name='pdbligand=GAL:BETA-D-GALACTOSE'>GAL</scene>, <scene name='pdbligand=NAG:N-ACETYL-D-GLUCOSAMINE'>NAG</scene></td></tr> | ||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=3zr6 FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=3zr6 OCA], [https://pdbe.org/3zr6 PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=3zr6 RCSB], [https://www.ebi.ac.uk/pdbsum/3zr6 PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=3zr6 ProSAT]</span></td></tr> | |||
</table> | |||
== Disease == | |||
[https://www.uniprot.org/uniprot/GALC_MOUSE GALC_MOUSE] Defects in Galc are the cause of the 'twitcher' phenotype; an autosomal recessive leukodystrophy similar to the human disease (Krabbe disease). This deficiency results in the insufficient catabolism of several galactolipids that are important in the production of normal myelin. | |||
== Function == | |||
[https://www.uniprot.org/uniprot/GALC_MOUSE GALC_MOUSE] Hydrolyzes the galactose ester bonds of galactosylceramide, galactosylsphingosine, lactosylceramide, and monogalactosyldiglyceride. Enzyme with very low activity responsible for the lysosomal catabolism of galactosylceramide, a major lipid in myelin, kidney and epithelial cells of small intestine and colon.<ref>PMID:8769874</ref> | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Krabbe disease is a devastating neurodegenerative disease characterized by widespread demyelination that is caused by defects in the enzyme galactocerebrosidase (GALC). Disease-causing mutations have been identified throughout the GALC gene. However, a molecular understanding of the effect of these mutations has been hampered by the lack of structural data for this enzyme. Here we present the crystal structures of GALC and the GALC-product complex, revealing a novel domain architecture with a previously uncharacterized lectin domain not observed in other hydrolases. All three domains of GALC contribute residues to the substrate-binding pocket, and disease-causing mutations are widely distributed throughout the protein. Our structures provide an essential insight into the diverse effects of pathogenic mutations on GALC function in human Krabbe variants and a compelling explanation for the severity of many mutations associated with fatal infantile disease. The localization of disease-associated mutations in the structure of GALC will facilitate identification of those patients that would be responsive to pharmacological chaperone therapies. Furthermore, our structure provides the atomic framework for the design of such drugs. | |||
Insights into Krabbe disease from structures of galactocerebrosidase.,Deane JE, Graham SC, Kim NN, Stein PE, McNair R, Cachon-Gonzalez MB, Cox TM, Read RJ Proc Natl Acad Sci U S A. 2011 Sep 13;108(37):15169-73. Epub 2011 Aug 29. PMID:21876145<ref>PMID:21876145</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 3zr6" style="background-color:#fffaf0;"></div> | |||
==See Also== | |||
*[[Galactosylceramidase|Galactosylceramidase]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
== | [[Category: Large Structures]] | ||
[[ | |||
== | |||
< | |||
[[Category: | |||
[[Category: Mus musculus]] | [[Category: Mus musculus]] | ||
[[Category: Cachon-Gonzalez | [[Category: Cachon-Gonzalez MB]] | ||
[[Category: Cox | [[Category: Cox TM]] | ||
[[Category: Deane | [[Category: Deane JE]] | ||
[[Category: Graham | [[Category: Graham SC]] | ||
[[Category: Kim | [[Category: Kim NN]] | ||
[[Category: Mcnair | [[Category: Mcnair R]] | ||
[[Category: Read | [[Category: Read RJ]] | ||
[[Category: Stein | [[Category: Stein PE]] | ||
Latest revision as of 14:10, 20 December 2023
STRUCTURE OF GALACTOCEREBROSIDASE FROM MOUSE IN COMPLEX WITH GALACTOSESTRUCTURE OF GALACTOCEREBROSIDASE FROM MOUSE IN COMPLEX WITH GALACTOSE
Structural highlights
DiseaseGALC_MOUSE Defects in Galc are the cause of the 'twitcher' phenotype; an autosomal recessive leukodystrophy similar to the human disease (Krabbe disease). This deficiency results in the insufficient catabolism of several galactolipids that are important in the production of normal myelin. FunctionGALC_MOUSE Hydrolyzes the galactose ester bonds of galactosylceramide, galactosylsphingosine, lactosylceramide, and monogalactosyldiglyceride. Enzyme with very low activity responsible for the lysosomal catabolism of galactosylceramide, a major lipid in myelin, kidney and epithelial cells of small intestine and colon.[1] Publication Abstract from PubMedKrabbe disease is a devastating neurodegenerative disease characterized by widespread demyelination that is caused by defects in the enzyme galactocerebrosidase (GALC). Disease-causing mutations have been identified throughout the GALC gene. However, a molecular understanding of the effect of these mutations has been hampered by the lack of structural data for this enzyme. Here we present the crystal structures of GALC and the GALC-product complex, revealing a novel domain architecture with a previously uncharacterized lectin domain not observed in other hydrolases. All three domains of GALC contribute residues to the substrate-binding pocket, and disease-causing mutations are widely distributed throughout the protein. Our structures provide an essential insight into the diverse effects of pathogenic mutations on GALC function in human Krabbe variants and a compelling explanation for the severity of many mutations associated with fatal infantile disease. The localization of disease-associated mutations in the structure of GALC will facilitate identification of those patients that would be responsive to pharmacological chaperone therapies. Furthermore, our structure provides the atomic framework for the design of such drugs. Insights into Krabbe disease from structures of galactocerebrosidase.,Deane JE, Graham SC, Kim NN, Stein PE, McNair R, Cachon-Gonzalez MB, Cox TM, Read RJ Proc Natl Acad Sci U S A. 2011 Sep 13;108(37):15169-73. Epub 2011 Aug 29. PMID:21876145[2] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||