4mzg: Difference between revisions
No edit summary |
No edit summary |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
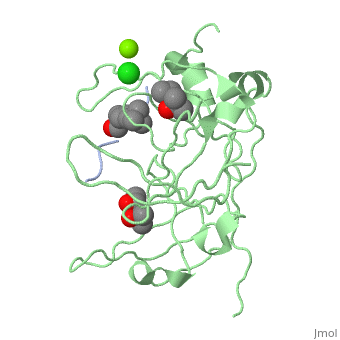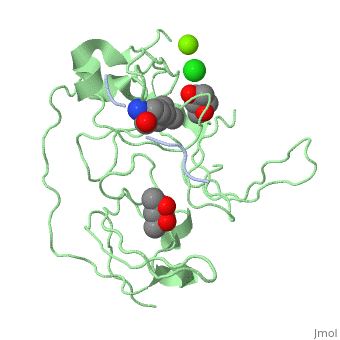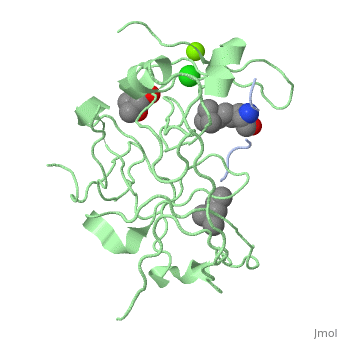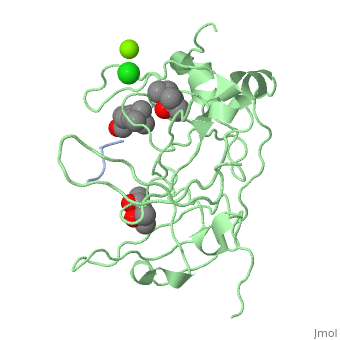
==Crystal structure of human Spindlin1 bound to histone H3K4me3 peptide== | |||
<StructureSection load='4mzg' size='340' side='right'caption='[[4mzg]], [[Resolution|resolution]] 1.70Å' scene=''> | |||
== Structural highlights == | |||
<table><tr><td colspan='2'>[[4mzg]] is a 4 chain structure with sequence from [https://en.wikipedia.org/wiki/Homo_sapiens Homo sapiens]. Full crystallographic information is available from [http://oca.weizmann.ac.il/oca-bin/ocashort?id=4MZG OCA]. For a <b>guided tour on the structure components</b> use [https://proteopedia.org/fgij/fg.htm?mol=4MZG FirstGlance]. <br> | |||
</td></tr><tr id='ligand'><td class="sblockLbl"><b>[[Ligand|Ligands:]]</b></td><td class="sblockDat" id="ligandDat"><scene name='pdbligand=CL:CHLORIDE+ION'>CL</scene>, <scene name='pdbligand=M3L:N-TRIMETHYLLYSINE'>M3L</scene>, <scene name='pdbligand=MG:MAGNESIUM+ION'>MG</scene>, <scene name='pdbligand=MPD:(4S)-2-METHYL-2,4-PENTANEDIOL'>MPD</scene>, <scene name='pdbligand=MRD:(4R)-2-METHYLPENTANE-2,4-DIOL'>MRD</scene></td></tr> | |||
<tr id='resources'><td class="sblockLbl"><b>Resources:</b></td><td class="sblockDat"><span class='plainlinks'>[https://proteopedia.org/fgij/fg.htm?mol=4mzg FirstGlance], [http://oca.weizmann.ac.il/oca-bin/ocaids?id=4mzg OCA], [https://pdbe.org/4mzg PDBe], [https://www.rcsb.org/pdb/explore.do?structureId=4mzg RCSB], [https://www.ebi.ac.uk/pdbsum/4mzg PDBsum], [https://prosat.h-its.org/prosat/prosatexe?pdbcode=4mzg ProSAT]</span></td></tr> | |||
</table> | |||
== Function == | |||
[https://www.uniprot.org/uniprot/H32_HUMAN H32_HUMAN] | |||
<div style="background-color:#fffaf0;"> | |||
== Publication Abstract from PubMed == | |||
Histone modification patterns and their combinatorial readout have emerged as a fundamental mechanism for epigenetic regulation. Here we characterized Spindlin1 as a histone effector that senses a cis-tail histone H3 methylation pattern involving trimethyllysine 4 (H3K4me3) and asymmetric dimethylarginine 8 (H3R8me2a) marks. Spindlin1 consists of triple tudor-like Spin/Ssty repeats. Cocrystal structure determination established concurrent recognition of H3K4me3 and H3R8me2a by Spin/Ssty repeats 2 and 1, respectively. Both H3K4me3 and H3R8me2a are recognized using an "insertion cavity" recognition mode, contributing to a methylation state-specific layer of regulation. In vivo functional studies suggest that Spindlin1 activates Wnt/beta-catenin signaling downstream from protein arginine methyltransferase 2 (PRMT2) and the MLL complex, which together are capable of generating a specific H3 "K4me3-R8me2a" pattern. Mutagenesis of Spindlin1 reader pockets impairs activation of Wnt target genes. Taken together, our work connects a histone "lysine-arginine" methylation pattern readout by Spindlin1-to-Wnt signaling at the transcriptional level. | |||
Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1.,Su X, Zhu G, Ding X, Lee SY, Dou Y, Zhu B, Wu W, Li H Genes Dev. 2014 Mar 15;28(6):622-36. doi: 10.1101/gad.233239.113. Epub 2014 Mar, 3. PMID:24589551<ref>PMID:24589551</ref> | |||
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.<br> | |||
</div> | |||
<div class="pdbe-citations 4mzg" style="background-color:#fffaf0;"></div> | |||
==See Also== | |||
*[[Spindlin|Spindlin]] | |||
== References == | |||
<references/> | |||
__TOC__ | |||
</StructureSection> | |||
[[Category: Homo sapiens]] | |||
[[Category: Large Structures]] | |||
[[Category: Ding X]] | |||
[[Category: Li H]] | |||
[[Category: Su X]] | |||
Latest revision as of 13:22, 28 December 2022
Crystal structure of human Spindlin1 bound to histone H3K4me3 peptideCrystal structure of human Spindlin1 bound to histone H3K4me3 peptide
Structural highlights
FunctionPublication Abstract from PubMedHistone modification patterns and their combinatorial readout have emerged as a fundamental mechanism for epigenetic regulation. Here we characterized Spindlin1 as a histone effector that senses a cis-tail histone H3 methylation pattern involving trimethyllysine 4 (H3K4me3) and asymmetric dimethylarginine 8 (H3R8me2a) marks. Spindlin1 consists of triple tudor-like Spin/Ssty repeats. Cocrystal structure determination established concurrent recognition of H3K4me3 and H3R8me2a by Spin/Ssty repeats 2 and 1, respectively. Both H3K4me3 and H3R8me2a are recognized using an "insertion cavity" recognition mode, contributing to a methylation state-specific layer of regulation. In vivo functional studies suggest that Spindlin1 activates Wnt/beta-catenin signaling downstream from protein arginine methyltransferase 2 (PRMT2) and the MLL complex, which together are capable of generating a specific H3 "K4me3-R8me2a" pattern. Mutagenesis of Spindlin1 reader pockets impairs activation of Wnt target genes. Taken together, our work connects a histone "lysine-arginine" methylation pattern readout by Spindlin1-to-Wnt signaling at the transcriptional level. Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1.,Su X, Zhu G, Ding X, Lee SY, Dou Y, Zhu B, Wu W, Li H Genes Dev. 2014 Mar 15;28(6):622-36. doi: 10.1101/gad.233239.113. Epub 2014 Mar, 3. PMID:24589551[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||