C-JUN: Difference between revisions
No edit summary |
Michal Harel (talk | contribs) No edit summary |
||
| (35 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
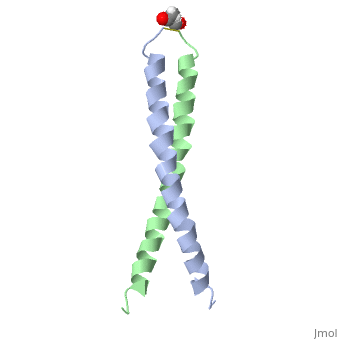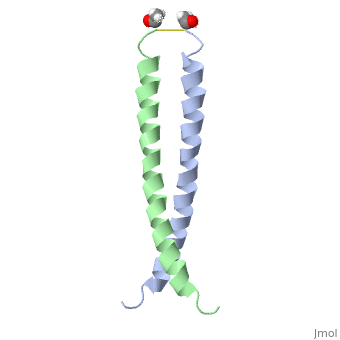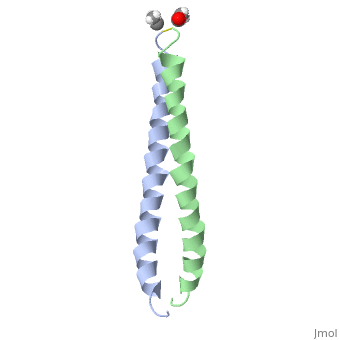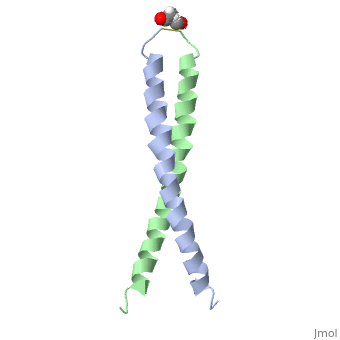
<StructureSection load='1jun' size='350' side='right' scene='' caption='Human C-Jun homodimer leucine zipper domain complex with acetyl (PDB code [[1jun]])'> | |||
== Introduction == | == Introduction == | ||
The c-Jun protein is a member of transcription factors which consist of a basic region leucine zipper region <ref name="one">PMID:8662824</ref>. Originally identified by its homology to v-jun, the oncogene from the avian sarcomoa virus <ref name="four"> Bossy-Wetzel, E., Bakiri, L., Yaniv, M. (1997). Induction of apoptosis by the transcription factor c-Jun. EMO Journal. Vol.16;7. 1695-1709 </ref>. All these leucine zipper factors bind to DNA in one of two states: homo or heterodimers <ref name="two">PMID:8662824</ref>. In conjunction with the c-Fos protein these two proteins bind to specific regions of DNA strands. Together these two proteins form the c-fos/c-jun complex which help regulate cell growth and differentiation <ref name="one">. | The '''c-Jun''' protein is a member of transcription factors which consist of a basic region leucine zipper region <ref name="one"> PMID:8662824 </ref>. Originally identified by its homology to v-jun, the oncogene from the avian sarcomoa virus <ref name="four"> Bossy-Wetzel, E., Bakiri, L., Yaniv, M. (1997). Induction of apoptosis by the transcription factor c-Jun. EMO Journal. Vol.16;7. 1695-1709 </ref>. All these leucine zipper factors bind to DNA in one of two states: homo or heterodimers <ref name="two"> PMID:8662824 </ref>. In conjunction with the c-Fos protein these two proteins bind to specific regions of DNA strands. Together these two proteins form the c-fos/c-jun complex which help regulate cell growth and differentiation <ref name="one" />. The members of the jun and fos families include three Jun proteins and four Fos proteins (c-Jun, JunB, JunD,c-Fos, Fos-B, Fra1, and Fra2) <ref name="one" />. Regulation of the complex iteslf is done by interactions between the protein and DNA in addition to the protein-protein interactions between each of the leucine zipper domains <ref name="one" />. See [[Transcription and RNA Processing]]. | ||
== Structure Overview == | == Structure Overview == | ||
The structure of c-Jun is comprised of a leucine zipper as previously stated. This dimerization motif may be in one of two classes, both of which are required for DNA-binding transcription factors; the basic-domain leucine zipper proteins (bZIP) and the basic helix loop-helix-leucine zipper proteins(bHLH-ZIP) <ref name="two"> A Junius, F.K., Mackay, J.P., Bubb, W.A., Jensen, S.A., Weiss, A.S., King, G.F. 2006. Nuclear Magnetic Resonance Characterization of the Jun Leucine Zipper Domain: Unusual Properties of Coiled-Coil Interfacial Polar Residues?</ref>. The strand becomes an elongated coiled coil. This is formed by residues at the a and d positions in each of the two monomers, whereby they create hydrophobic centers which conform to the "knobs into holes" model by Crick. <ref name="two" />. Amino acids at these a and d positions are each surrounded by 4 additional residues from adjacent a-helix monomer <ref name="two" />. | |||
The a and d residues each exhibit varying types of packing in terms of this "knobs into holes" theory. According to Harbury et al.(24) the leucines at the a positions are packed "parallel" in such a way that the C-alpha-C-beta bond vector lies in a parallel manner to the C-alpha-C-alpha vector at the base of the acceptor hole on adjacent helix <ref name="one" />. Whereas the opposite is true for the leucines in the d positions. Here the residues are packed in a "perpendicular" nature <ref name="one" />. The bond vector of the C-alpha-C-beta pack approximately perpendicular to the C-alpha-C-alpha vector at the base of the hole of the second helix in which it packs <ref name="one" />. Therefore only the leucine side chains in the a positions, which point away from the boundary, make van der Waals interactions <ref name="one" />. | |||
== Protein Function == | == Protein Function == | ||
The primary function of c-Jun is in regards to DNA transcription. Specifically, the protein is involved in proliferation, apoptosis, oncogenic transformation and various cellular processes <ref name="three">PMID:12798298</ref>. | The primary function of c-Jun is in regards to DNA transcription. Specifically, the protein is involved in proliferation, apoptosis, oncogenic transformation and various cellular processes <ref name="three"> PMID:12798298 </ref>. For instance cells which lack an allele for c-jun have been shown to stunt growth both in vitro and in vivo <ref name="four" />. Whereas a prolonged and therefore strong induction of c-jun has been in response to such things as tumor necrosis factor or stress inducing stimuli such as ultra violet radiation <ref name="four" />. | ||
== Protein Regulation == | == Protein Regulation == | ||
Changes made in the phosphorylation state of specific amino acids is one means by which c-Jun regulates transcription <ref name=" | Changes made in the phosphorylation state of specific amino acids is one means by which c-Jun regulates transcription <ref name="six"> PMID:8165146 </ref>. To date two seperate sites of phosphorylation have been identified. One is located at the N-terminal end in which the amino acids Ser63 and Ser73 are phosphorylated in response to ''ras'' expression. When ''ras'' is expressed, and Ser63 and Ser73 are phosphorylated,and transcriptional activity of c-Jun increases. The second site is located at the C-terminal which is very close in proximity to the DNA binding domain. Here the residues are Thr214, Ser226, and Ser 232 <ref name="six" />. Unlike the two serines at the N-terminal end, phosphorylation at the C-terminal end inhibits DNA binding to c-Jun <ref name="six" />. Therefore with the expression of such oncogenes as ''ras'' dephsphorylation of these three residues occurs. | ||
== Psychological Influences == | == Psychological Influences == | ||
The stress-induced | The stress-induced signaling cascade may also active c-Jun by phosphorylation. The N-ternminal protein kinase phosphorylates Ser63 and Ser73 <ref name="five"> PMID:10064599 </ref> . Another mechanism for the activation however is interestingly through intracellular calcium concentrations. Increasing these concentrations by opening the L-type voltage gated calcium channels leads to serines phosphorlation. | ||
It was found that the N-terminus contains both calcium and stress-regulated transcriptional activation domains <ref name=" | It was found that the N-terminus contains both calcium and stress-regulated transcriptional activation domains <ref name="five" />. According to the study,distinct mechanisms of c-Jun control function by calcium and stress signals <ref name="five" />. | ||
==Additional Resources== | |||
To See Additional information, see: [[Transcription and RNA Processing]] <br /> | |||
</StructureSection> | |||
==3D structure of C-JUN== | |||
Updated on {{REVISIONDAY2}}-{{MONTHNAME|{{REVISIONMONTH}}}}-{{REVISIONYEAR}} | |||
[[1jun]] – hCJUN leucine zipper domain – human – NMR<br /> | |||
[[1jnm]] - hCJUN leucine zipper domain + DNA<br /> | |||
[[1fos]] – hCJUN + p55 c-Fos + DNA<br /> | |||
[[5fv8]] – hCJUN + FOSW<br /> | |||
== References == | == References == | ||
<references/> | <references/> | ||
[[Category:Topic Page]] | |||
Latest revision as of 12:14, 11 April 2018
IntroductionThe c-Jun protein is a member of transcription factors which consist of a basic region leucine zipper region [1]. Originally identified by its homology to v-jun, the oncogene from the avian sarcomoa virus [2]. All these leucine zipper factors bind to DNA in one of two states: homo or heterodimers [3]. In conjunction with the c-Fos protein these two proteins bind to specific regions of DNA strands. Together these two proteins form the c-fos/c-jun complex which help regulate cell growth and differentiation [1]. The members of the jun and fos families include three Jun proteins and four Fos proteins (c-Jun, JunB, JunD,c-Fos, Fos-B, Fra1, and Fra2) [1]. Regulation of the complex iteslf is done by interactions between the protein and DNA in addition to the protein-protein interactions between each of the leucine zipper domains [1]. See Transcription and RNA Processing. Structure OverviewThe structure of c-Jun is comprised of a leucine zipper as previously stated. This dimerization motif may be in one of two classes, both of which are required for DNA-binding transcription factors; the basic-domain leucine zipper proteins (bZIP) and the basic helix loop-helix-leucine zipper proteins(bHLH-ZIP) [3]. The strand becomes an elongated coiled coil. This is formed by residues at the a and d positions in each of the two monomers, whereby they create hydrophobic centers which conform to the "knobs into holes" model by Crick. [3]. Amino acids at these a and d positions are each surrounded by 4 additional residues from adjacent a-helix monomer [3]. The a and d residues each exhibit varying types of packing in terms of this "knobs into holes" theory. According to Harbury et al.(24) the leucines at the a positions are packed "parallel" in such a way that the C-alpha-C-beta bond vector lies in a parallel manner to the C-alpha-C-alpha vector at the base of the acceptor hole on adjacent helix [1]. Whereas the opposite is true for the leucines in the d positions. Here the residues are packed in a "perpendicular" nature [1]. The bond vector of the C-alpha-C-beta pack approximately perpendicular to the C-alpha-C-alpha vector at the base of the hole of the second helix in which it packs [1]. Therefore only the leucine side chains in the a positions, which point away from the boundary, make van der Waals interactions [1].
Protein FunctionThe primary function of c-Jun is in regards to DNA transcription. Specifically, the protein is involved in proliferation, apoptosis, oncogenic transformation and various cellular processes [4]. For instance cells which lack an allele for c-jun have been shown to stunt growth both in vitro and in vivo [2]. Whereas a prolonged and therefore strong induction of c-jun has been in response to such things as tumor necrosis factor or stress inducing stimuli such as ultra violet radiation [2]. Protein RegulationChanges made in the phosphorylation state of specific amino acids is one means by which c-Jun regulates transcription [5]. To date two seperate sites of phosphorylation have been identified. One is located at the N-terminal end in which the amino acids Ser63 and Ser73 are phosphorylated in response to ras expression. When ras is expressed, and Ser63 and Ser73 are phosphorylated,and transcriptional activity of c-Jun increases. The second site is located at the C-terminal which is very close in proximity to the DNA binding domain. Here the residues are Thr214, Ser226, and Ser 232 [5]. Unlike the two serines at the N-terminal end, phosphorylation at the C-terminal end inhibits DNA binding to c-Jun [5]. Therefore with the expression of such oncogenes as ras dephsphorylation of these three residues occurs. Psychological InfluencesThe stress-induced signaling cascade may also active c-Jun by phosphorylation. The N-ternminal protein kinase phosphorylates Ser63 and Ser73 [6] . Another mechanism for the activation however is interestingly through intracellular calcium concentrations. Increasing these concentrations by opening the L-type voltage gated calcium channels leads to serines phosphorlation. It was found that the N-terminus contains both calcium and stress-regulated transcriptional activation domains [6]. According to the study,distinct mechanisms of c-Jun control function by calcium and stress signals [6]. Additional ResourcesTo See Additional information, see: Transcription and RNA Processing |
| ||||||||||
3D structure of C-JUN3D structure of C-JUN
Updated on 11-April-2018
1jun – hCJUN leucine zipper domain – human – NMR
1jnm - hCJUN leucine zipper domain + DNA
1fos – hCJUN + p55 c-Fos + DNA
5fv8 – hCJUN + FOSW
ReferencesReferences
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 1.7 Junius FK, O'Donoghue SI, Nilges M, Weiss AS, King GF. High resolution NMR solution structure of the leucine zipper domain of the c-Jun homodimer. J Biol Chem. 1996 Jun 7;271(23):13663-7. PMID:8662824
- ↑ 2.0 2.1 2.2 Bossy-Wetzel, E., Bakiri, L., Yaniv, M. (1997). Induction of apoptosis by the transcription factor c-Jun. EMO Journal. Vol.16;7. 1695-1709
- ↑ 3.0 3.1 3.2 3.3 Junius FK, O'Donoghue SI, Nilges M, Weiss AS, King GF. High resolution NMR solution structure of the leucine zipper domain of the c-Jun homodimer. J Biol Chem. 1996 Jun 7;271(23):13663-7. PMID:8662824 Cite error: Invalid
<ref>tag; name "two" defined multiple times with different content - ↑ Mechta-Grigoriou F, Giudicelli F, Pujades C, Charnay P, Yaniv M. c-jun regulation and function in the developing hindbrain. Dev Biol. 2003 Jun 15;258(2):419-31. PMID:12798298
- ↑ 5.0 5.1 5.2 Hoeffler WK, Levinson AD, Bauer EA. Activation of c-Jun transcription factor by substitution of a charged residue in its N-terminal domain. Nucleic Acids Res. 1994 Apr 11;22(7):1305-12. PMID:8165146
- ↑ 6.0 6.1 6.2 Cruzalegui FH, Hardingham GE, Bading H. c-Jun functions as a calcium-regulated transcriptional activator in the absence of JNK/SAPK1 activation. EMBO J. 1999 Mar 1;18(5):1335-44. PMID:10064599 doi:10.1093/emboj/18.5.1335