User:Alice Harmon/EF Hand: Difference between revisions
Alice Harmon (talk | contribs) No edit summary |
Alice Harmon (talk | contribs) No edit summary |
||
| (8 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
EF-hands are calcium-binding motifs found in hundreds of proteins. They bind calcium ions with high affinity (K<sub>d</sub>s are in the micromolar range) and selectivity, and this property allows EF | [[Image:EF-hand.jpg|left]] | ||
EF hands are calcium-binding motifs found in hundreds of proteins. They bind calcium ions with high affinity (K<sub>d</sub>s are in the micromolar range) and selectivity, and this property allows EF hand proteins to sense changes in intracellular calcium. In unstimulated cells cellular free calcium concentrations [Ca<sup>2+</sup>]<sub>c</sub> are in the nanomolar range (~10 nM in animal cells and ~200 nM in plant cells), and EF hands are generally unoccupied by Ca<sup>2+</sup>. Upon stimulation, Ca<sup>2+</sup> enters the cytosol from either outside the cell or from internal organelles, and [Ca<sup>2+</sup>]<sub>c</sub> rises to the micromolar range. EF hands bind Ca<sup>2+</sup>, and this binding causes a conformational change that alters the activity of the protein. | |||
The name EF hand originated from the first such structure to be described, which was in the protein [[parvalbumin]]<ref>PMID:4700463</ref>. In this protein calcium is bound by a helix-loop-helix structure that is formed by the E and F helices (letters assigned to helices in the order that they occur, starting at the N-terminus). See the annotated protein sequence for carp parvalbumin here [http://www.pdb.org/pdb/explore/remediatedSequence.do?structureId=4CPV&bionumber=1]. The structure resembles a hand with the forefinger pointing in the direction of the E helix, the thumb pointing in the direction of the H helix, and the remaining fingers curled to resemble the calcium-binding loop. | |||
The | The loop structure is formed by the EF hand calcium-binding motif, which contains 12 residues and is defined in Prosite concensus pattern PS00018 <ref>http://prosite.expasy.org/PDOC00018</ref> <br> | ||
{| | {| | ||
|Residue | |Residue: | ||
|D-x-[DNS]-{ILVFYW}-[DENSTG]-[DNQGHRK]-{GP}-[LIVMC]-[DENQSTAGC]- x(2) -[DE]<br> | |<font face = courier>D-x-[DNS]-{ILVFYW}-[DENSTG]-[DNQGHRK]-{GP}-[LIVMC]-[DENQSTAGC]- x(2) -[DE]</font><br> | ||
|- | |- | ||
|Position | |Position: | ||
|1-2-3< | |<font face =courier>1-2-<font color='white'>--</font>3<font color='white'>--</font>-<font color='white'>---</font>4<font color='white'>xxxx</font>-<font color='white'>---</font>5<font color='white'>----</font>-<font color='white'>----</font>6<font color='white'>----</font>-<font color='white'>-</font>7<font color='white'>--</font>-<font color='white'>--</font>8<font color='white'>----</font>-<font color='white'>----</font>9<font color='white'>------</font>-10,11<font color='white'>-</font>-<font color='white'>-</font>12 | ||
|} | |} | ||
where x indicates any residue; any residue in square brackets [ ] is possible at that position; none of the residues in curly brackets { } are possible: and x(2) indicates a series of two x's. | |||
[[Image:Pyramid small.jpg|right]] | |||
The calcium ion is coordinated by seven oxygen atoms that form a pentagonal bipyramid as shown in the figure to the right. One oxygen is contributed by the side chain of each residue in positions 1, 3, 5, and 7 of the motif; two are from the side chain of E/D at position 12; one is from the backbone of the residue at position 7; and one from water. Labels in the figure indicate the arrangement of the oxygen atoms in the structure. | |||
The 3D structure of EF hands is depicted in the scenes below, which show three examples of EF hand proteins. [[Parvalbumin]] is a monomeric protein that has a pair of EF hands, [[calmodulin]] is a monomer with two pairs, and [[Calcium-dependent protein kinase]] is a monomer with a protein kinase catalytic domain and its calcium-binding domain has two pairs of EF hands. As shown in these examples EF hands often occur in interacting pairs, which enables the cooperative binding of calcium ions. | |||
Scenes numbered 1 are the default scenes showing the proteins in cartoon with Ca<sup>2+</sup> in green space fill. The calcium binding domain of CDPK is blue and the kinase catalytic domain is in gold and has an ATP analog (sticks in CPK colors) bound in its active site.<br> | |||
Scenes numbered 2 show pairs of EF hands in each protein, one in blue and one in gold, which are linked by the sequence shown in orchid.<br> | |||
Scenes numbered 3 isolate one EF hand: the eponymous EF hand of parvalbumen, EF hand I (they are numbered I-IV) in calmodulin, and EF hand IV of CDPK.<br> | |||
Scenes numbered 4 show the calcium binding loops in the same orientation. The protein backbone is shown as a trace and the sidechains of residues that provide ligands are shown in ball and stick. The five oxygen atoms that form the the base of the bipyramid are contributed by four residues (positions 3, 5, 7, and 12) distributed around the equator of the calcium ion, one pyramid point ("north" in the scenes) is the oxygen from water, and the "south" point is the side chain oxygen from the invariant D at position 1 in the motif. | |||
{| | {| | ||
| <applet load='4cpv' size='300' frame='true' align='right' caption='4cpv - Carp parvalbumin' scene='56/562354/Parvalbumin/1' /><Br>'''4cpv''' <Br><scene name='56/562354/Parvalbumin/2'> | | <applet load='4cpv' size='300' frame='true' align='right' caption='4cpv - Carp parvalbumin' scene='56/562354/Parvalbumin/1' /><Br>'''4cpv''' <Br><scene name='56/562354/Parvalbumin/1'>1. Holoprotein</scene><Br><scene name='56/562354/Parvalbumin/2'>2. Pair of EF hands</scene><br><scene name='56/562354/Parvalbumin/3'>3. The original EF-Hand</scene><br><scene name='56/562354/Parvalbumin/6'>4. Calcium-binding loop</scene> | ||
| <applet load='1prw' size='300' frame='true' align='right' caption='1prw - Bovine calmodulin' scene='56/562354/Calmodulin/1' /><Br>'''1prw''' <Br><scene name='56/562354/Calmodulin/3'> | | <applet load='1prw' size='300' frame='true' align='right' caption='1prw - Bovine calmodulin' scene='56/562354/Calmodulin/1' /><Br>'''1prw''' <Br><scene name='56/562354/Calmodulin/1'>1. Holoprotein</scene><Br><scene name='56/562354/Calmodulin/3'>2. Pair of EF hands</scene><br><scene name='56/562354/Calmodulin/8'>2. EF hand I</scene><br><scene name='56/562354/Calmodulin/7'>4. Calcium-binding loop</scene> | ||
| <applet load='3HX4' size='300' frame='true' align='left' caption='3hx4 - active TgCDPK1' scene = '56/562354/Cdpk/1' /><Br>'''3HX4'''<Br><scene name='56/562354/Cdpk_ef_hand_pair/2'> | | <applet load='3HX4' size='300' frame='true' align='left' caption='3hx4 - active TgCDPK1' scene = '56/562354/Cdpk/1' /><Br>'''3HX4'''<Br><scene name='56/562354/Cdpk/1'>1. Holoprotein</scene><Br><scene name='56/562354/Cdpk_ef_hand_pair/2'>2. Pair of EF hands</scene><br><scene name='56/562354/Cdpk_ef_iv/1'>3. EF hand IV</scene><br><scene name='56/562354/Cdpk_ef_iv/3'>4. Calcium-binding loop</scene> | ||
|} | |} | ||
=References= | |||
<references/> | |||
Latest revision as of 16:28, 24 September 2013
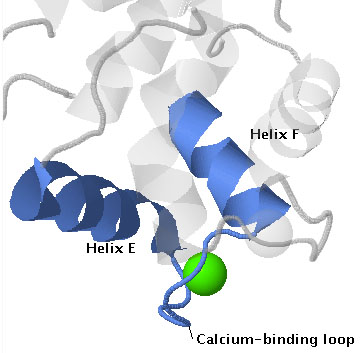
EF hands are calcium-binding motifs found in hundreds of proteins. They bind calcium ions with high affinity (Kds are in the micromolar range) and selectivity, and this property allows EF hand proteins to sense changes in intracellular calcium. In unstimulated cells cellular free calcium concentrations [Ca2+]c are in the nanomolar range (~10 nM in animal cells and ~200 nM in plant cells), and EF hands are generally unoccupied by Ca2+. Upon stimulation, Ca2+ enters the cytosol from either outside the cell or from internal organelles, and [Ca2+]c rises to the micromolar range. EF hands bind Ca2+, and this binding causes a conformational change that alters the activity of the protein.
The name EF hand originated from the first such structure to be described, which was in the protein parvalbumin[1]. In this protein calcium is bound by a helix-loop-helix structure that is formed by the E and F helices (letters assigned to helices in the order that they occur, starting at the N-terminus). See the annotated protein sequence for carp parvalbumin here [1]. The structure resembles a hand with the forefinger pointing in the direction of the E helix, the thumb pointing in the direction of the H helix, and the remaining fingers curled to resemble the calcium-binding loop.
The loop structure is formed by the EF hand calcium-binding motif, which contains 12 residues and is defined in Prosite concensus pattern PS00018 [2]
| Residue: | D-x-[DNS]-{ILVFYW}-[DENSTG]-[DNQGHRK]-{GP}-[LIVMC]-[DENQSTAGC]- x(2) -[DE] |
| Position: | 1-2---3------4xxxx----5---------6------7-----8---------9-------10,11---12 |
where x indicates any residue; any residue in square brackets [ ] is possible at that position; none of the residues in curly brackets { } are possible: and x(2) indicates a series of two x's.
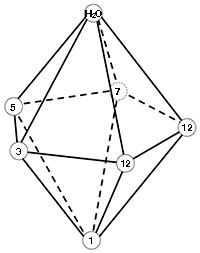
The calcium ion is coordinated by seven oxygen atoms that form a pentagonal bipyramid as shown in the figure to the right. One oxygen is contributed by the side chain of each residue in positions 1, 3, 5, and 7 of the motif; two are from the side chain of E/D at position 12; one is from the backbone of the residue at position 7; and one from water. Labels in the figure indicate the arrangement of the oxygen atoms in the structure.
The 3D structure of EF hands is depicted in the scenes below, which show three examples of EF hand proteins. Parvalbumin is a monomeric protein that has a pair of EF hands, calmodulin is a monomer with two pairs, and Calcium-dependent protein kinase is a monomer with a protein kinase catalytic domain and its calcium-binding domain has two pairs of EF hands. As shown in these examples EF hands often occur in interacting pairs, which enables the cooperative binding of calcium ions.
Scenes numbered 1 are the default scenes showing the proteins in cartoon with Ca2+ in green space fill. The calcium binding domain of CDPK is blue and the kinase catalytic domain is in gold and has an ATP analog (sticks in CPK colors) bound in its active site.
Scenes numbered 2 show pairs of EF hands in each protein, one in blue and one in gold, which are linked by the sequence shown in orchid.
Scenes numbered 3 isolate one EF hand: the eponymous EF hand of parvalbumen, EF hand I (they are numbered I-IV) in calmodulin, and EF hand IV of CDPK.
Scenes numbered 4 show the calcium binding loops in the same orientation. The protein backbone is shown as a trace and the sidechains of residues that provide ligands are shown in ball and stick. The five oxygen atoms that form the the base of the bipyramid are contributed by four residues (positions 3, 5, 7, and 12) distributed around the equator of the calcium ion, one pyramid point ("north" in the scenes) is the oxygen from water, and the "south" point is the side chain oxygen from the invariant D at position 1 in the motif.
4cpv |
1prw |
3HX4 |
ReferencesReferences
- ↑ Kretsinger RH, Nockolds CE. Carp muscle calcium-binding protein. II. Structure determination and general description. J Biol Chem. 1973 May 10;248(9):3313-26. PMID:4700463
- ↑ http://prosite.expasy.org/PDOC00018