Sandbox Reserved 554
| This Sandbox is Reserved from 05/22/2012, through 07/22/2012 for use in the course "BIOL 414" taught by Greg Buhrman at the North Carolina State University, Raleigh, NC USA. This reservation includes Sandbox Reserved 551 through Sandbox Reserved 590. |
To get started:
More help: Help:Editing |
Hi Lisa,
Here is your very own Protopedia page for pdb code: 3MJG. Your presentation date is scheduled for: June 20.
Have Fun!
Greg Buhrman

| |||||||
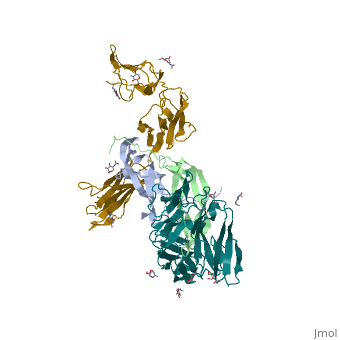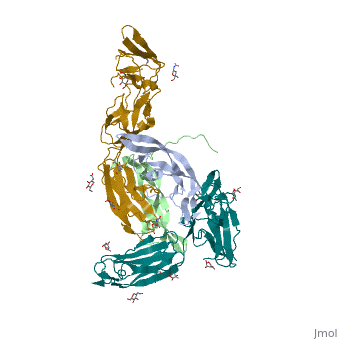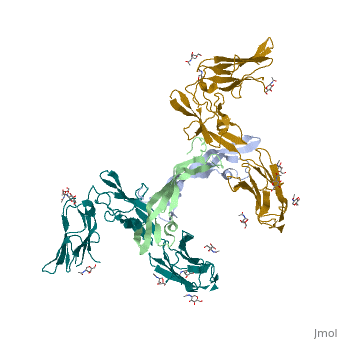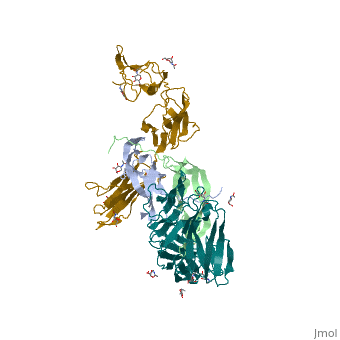
| 3mjg, resolution 2.30Å () | |||||||
|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||
| Gene: | PDGF2, PDGFB, SIS (Homo sapiens), PDGFRB (Homo sapiens) | ||||||
| Activity: | Receptor protein-tyrosine kinase, with EC number 2.7.10.1 | ||||||
| |||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||
The structure of a platelet derived growth factor receptor complexThe structure of a platelet derived growth factor receptor complex
Platelet-derived growth factors (PDGFs) and their receptors (PDGFRs) are prototypic growth factors and receptor tyrosine kinases which have critical functions in development. We show that PDGFs share a conserved region in their prodomain sequences which can remain noncovalently associated with the mature cystine-knot growth factor domain after processing. The structure of the PDGF-A/propeptide complex reveals this conserved, hydrophobic association mode. We also present the structure of the complex between PDGF-B and the first three Ig domains of PDGFRbeta, showing that two PDGF-B protomers clamp PDGFRbeta at their dimerization seam. The PDGF-B:PDGFRbeta interface is predominantly hydrophobic, and PDGFRs and the PDGF propeptides occupy overlapping positions on mature PDGFs, rationalizing the need of propeptides by PDGFs to cover functionally important hydrophobic surfaces during secretion. A large-scale structural organization and rearrangement is observed for PDGF-B upon receptor binding, in which the PDGF-B L1 loop, disordered in the structure of the free form, adopts a highly specific conformation to form hydrophobic interactions with the third Ig domain of PDGFRbeta. Calorimetric data also shows that the membrane-proximal homotypic PDGFRalpha interaction, albeit required for activation, contributes negatively to ligand binding. The structural and biochemical data together offer insights into PDGF-PDGFR signaling, as well as strategies for PDGF-antagonism.
Structures of a platelet-derived growth factor/propeptide complex and a platelet-derived growth factor/receptor complex., Shim AH, Liu H, Focia PJ, Chen X, Lin PC, He X, Proc Natl Acad Sci U S A. 2010 Jun 22;107(25):11307-12. Epub 2010 Jun 2. PMID:20534510
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
About this StructureAbout this Structure
3MJG is a 4 chains structure with sequences from Homo sapiens. Full crystallographic information is available from OCA.
ReferenceReference
Page seeded by OCA on Wed Aug 18 11:07:32 2010