Sandbox Reserved 100
Monomers Per TurnMonomers Per Turn
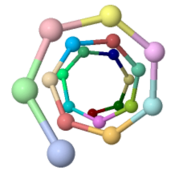
The ARC-1 model has 6.4 monomer chains per turn (56.0 degrees rotation between monomers). This is more chains/turn than some previous type IV pilus models.
To visualize chains/turn, we show (alpha carbon of Phe51). Then these chain-marking-atoms are , and the resulting helix is viewed from one end. When counting the chains/turn, bear in mind that the first and last (to complete one turn) count as 1/2 chain each.
|
Pilus |
Chains/Turn (Angle) |
Image |
|
Geobacter sulfurreducens (type IVa, 61 amino acids, theoretical model, 2016)[1] |
6.4 (56.0°) | |
|
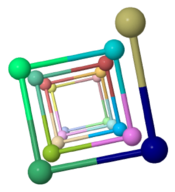
Pseudomonas aeruginosa (type IVa, 150 amino acids, fiber diffraction, 2004)[2] |
4.0 (90°) | |
|
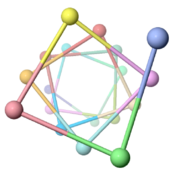
Vibrio cholerae (type IVb, 198 amino acids, cryo-EM, 2012)[3] |
3.7 (96.8°) | |
|
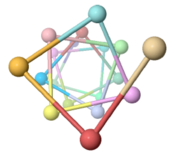
Neisseria gonorrhoeae (type IVa, 165 amino acids, cryo-EM, 2006)[4] |
3.6 (100.8°) |
Notes & ReferencesNotes & References
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedxiao1 - ↑ Craig L, Pique ME, Tainer JA. Type IV pilus structure and bacterial pathogenicity. Nat Rev Microbiol. 2004 May;2(5):363-78. PMID:15100690 doi:http://dx.doi.org/10.1038/nrmicro885
- ↑ Li J, Egelman EH, Craig L. Structure of the Vibrio cholerae Type IVb Pilus and stability comparison with the Neisseria gonorrhoeae type IVa pilus. J Mol Biol. 2012 Apr 20;418(1-2):47-64. doi: 10.1016/j.jmb.2012.02.017. Epub 2012, Feb 21. PMID:22361030 doi:http://dx.doi.org/10.1016/j.jmb.2012.02.017
- ↑ Craig L, Volkmann N, Arvai AS, Pique ME, Yeager M, Egelman EH, Tainer JA. Type IV pilus structure by cryo-electron microscopy and crystallography: implications for pilus assembly and functions. Mol Cell. 2006 Sep 1;23(5):651-62. PMID:16949362 doi:10.1016/j.molcel.2006.07.004