The Mechanism of TrypsinThe Mechanism of Trypsin
|
|
| 2ah4, resolution 1.13Å ()
|
| Ligands:
|
, ,
|
| Activity:
|
Trypsin, with EC number 3.4.21.4
|
| Related:
|
2age, 2agg, 2agi
|
|
|
|
|
|
|
| Resources:
|
FirstGlance, OCA, PDBsum, RCSB
|
| Coordinates:
|
save as pdb, mmCIF, xml
|
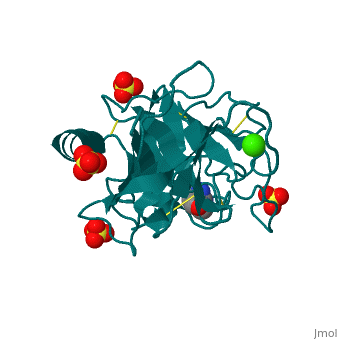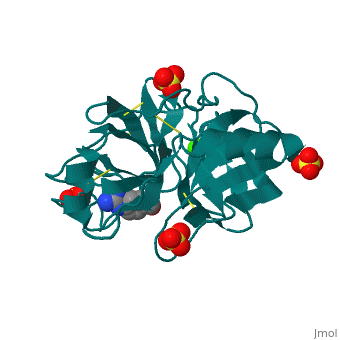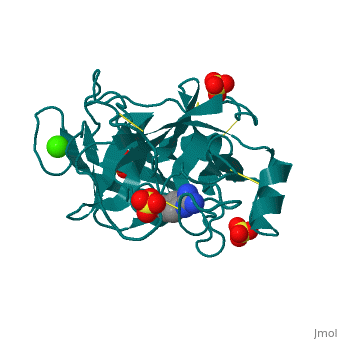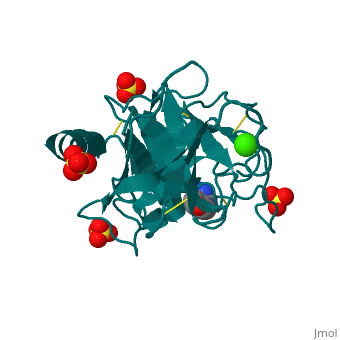
The secondary structure of trypsin (pdb code 2ah4) is composed of 13 antiparallel beta sheets with three alpha helices interspersed throughout. The sheets are divided into two groups—one with six sheets, and one with seven—each forming a tight barrel that appears relatively open on each end [1]. The three members of the catalytic triad in other serine proteases—His 57, Asp 102, and Ser 195—are located near one end of the group of six beta sheets “capped” by an alpha helix. This is logical, as the three work together to stabilize the scissile bond of residues with small side chains. His 57 and Asp 102, which both serve to stabilize via hydrogen bonding and as proton acceptors, lie outside the barrel proper. Serine 195, which forms the tetrahedral intermediate and “hydrolyzes” the peptide bond, also formally lies outside this barrel, but is placed such that it is able to easily interact with any peptide chain which passes through either barrel. Although it seems difficult to imagine how a peptide chain could fit within the relatively small barrels, the ability of certain “flaps” to move and open up the barrels—as in HIV protease—provides a possible explanation for this problem.
- ↑ Radisky ES, Lee JM, Lu CJ, Koshland DE Jr. Insights into the serine protease mechanism from atomic resolution structures of trypsin reaction intermediates. Proc Natl Acad Sci U S A. 2006 May 2;103(18):6835-40. Epub 2006 Apr 24. PMID:16636277
Proteopedia Page Contributors and Editors (what is this?)Proteopedia Page Contributors and Editors (what is this?)
Michael Vick