Michael Pikaart/Biochem StructureIntro
 Chem311 Course Page 1 Introduction to protein structure
Chem311 Course Page 1 Introduction to protein structure
IntroductionIntroduction
A protein's function depends on the proper folding of the linear polymeric chain of amino acids making up its polypeptide sequence. Folding of a polypeptide occurs by way of rotation of the single bonds in either side of each amino acid residue's alpha carbon, characterized by the Φ and Ψ angles. In folded proteins, stretches of amino acids typically have a re-iterated and restricted set of Φ and Ψ angles, giving rise to a length of stable secondary structure. If this structure has a rotational displacement, the result is a helical secondary structure; if the structure occurs in a zig-zag arrangement, a sheet secondary structure results. Here we will explore these two types of secondary structures, as found in actual proteins.
Alpha helixAlpha helix
Helix or screw structures are found in lots of contexts, both in nature and in man-made objects like these:
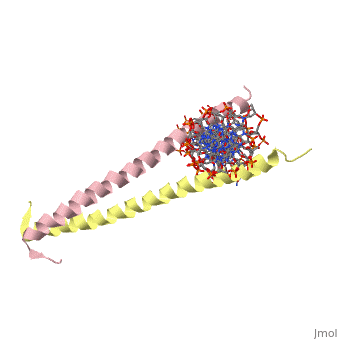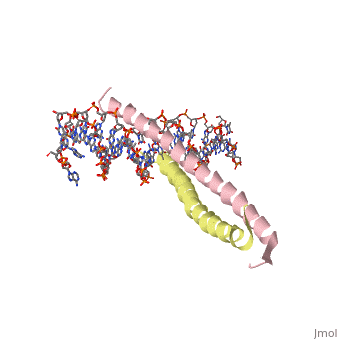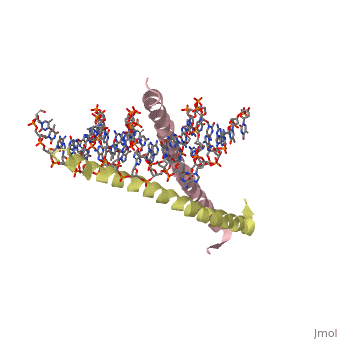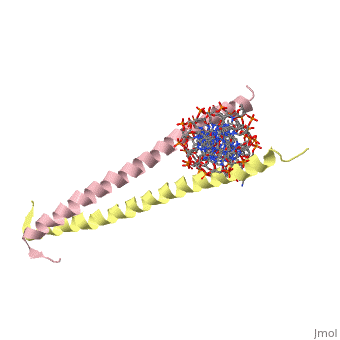
Helices have a twist to them, either right handed or left handed. A normal right-tighty screw is a right-handed helix. We take a look at a protein called GCN4, a transcription factor found in yeast that binds DNA and regulates expression of genes responsible for synthesis of amino acids. (For more information, go to [1]). The GCN4 protein has a long section of alpha helix. Part of this helix binds directly to a particular sequence of DNA, while the part of the helix not in direct contact with the DNA binds to a second GCN4 molecule. In fact, GCN4 must bind to a second GCN4 molecule in order to bind DNA; GCN4 thus binds DNA as a homodimer. Thus the alpha helix of GCN4 provides both its DNA binding domain and its dimerization domain. We'll start with looking at the GCN4 dimer bound to DNA. It looks kind of like you're picking up a pencil between two fingers. Take a minute to play around with this structure. By grabbing the image with a left-click, you can rotate it around. If you hold the Shift key as you left-click, you can either zoom, by moving your mouse up and down, or rotate in the z axis, by moving your mouse left and right. Right-clicking gives you a bunch of options allowing you to control how the structure appears. Then click the green links below to walk through a number of close-ups of the molecule.
Let's take a closer look at a bit of the alpha helix.
|
Zoom in and out a bit and make sure you can distinguish the backbone atoms from the side chains. See if you can tell by inspection what amino acids are present in this bit of helix. The green dashed lines show the i to i+4th residue hydrogen bonds. If you rotate the helix so you can see right down the helical axis, you would be lead to believe that there is quite a bit of space down the middle. To convince yourself that the helix is actually very tightly packed, left-click on the structure, chose "Style," then pick "Scheme," and then "CPK Spacefill."
This scene shows you the region of the dimerized proteins where they contact each other. The hydrophobic amino acid side chains leucine and valine are shown in green. Note how the amphipathic character of each helix. However, there is one amino acid side chain at the dimer interface which is not hydrophobic - can you tell which it is?
Here is a look at one of the GCN4 polypeptides in contact with DNA. Two side chains in proximity to the DNA are shown as balls, the DNA in stick form.
Beta sheetBeta sheet
A second type of structure commonly found in proteins is the beta sheet. Unlike the helix, there is not a rotational component to this structure. Because of this, a single length of beta sheet structure cannot complete intra-strand hydrogen bonding. Rather, the hydrogen bonding that stabilizes beta sheet structures occurs between neighboring strands. These strands can either be parallel or antiparallel. In either orientation, there must be a stretch of linker polypeptide in a continuous polypeptide - this may be as short as two amino acids in a so-called beta turn, or a longer stretch of polypeptide with its own secondary structure. A protein which has significant beta sheet secondary structure is the protease elastase:
|
: Here the beta sheet strands of elastase are highlighted. You can see the emergence of the higher-order domains formed within the protein's structure.
: This is a close-up look at classical beta turn in the enzyme carbonic anhydrase. Inter-strand hydrogen bonds are shown in green. See if you can identify the two amino acid residues at the tip of the beta turn.