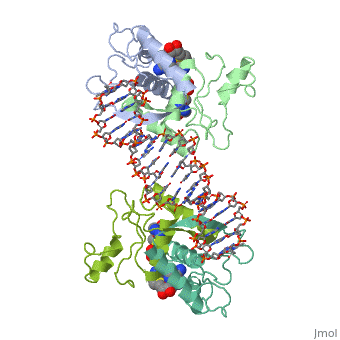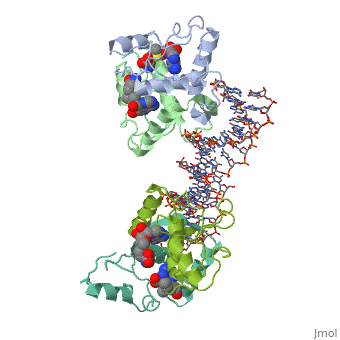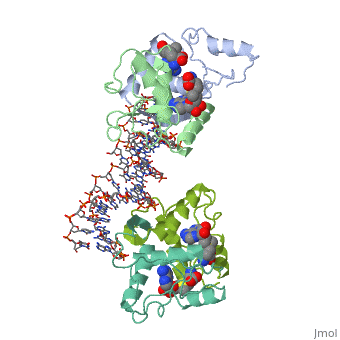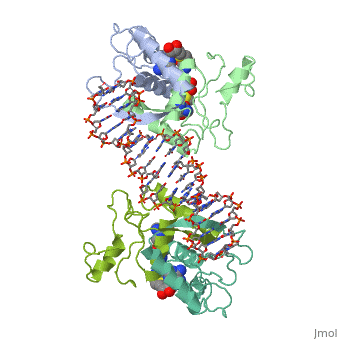
Met repressor sandbox

| |||||||||
| 1cma, resolution 2.80Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | |||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
E. coli met repressorE. coli met repressor
The regulates the transcription genes involved in the biosythesis of methionine in E. coli, and is the product of the metJ gene [1] Methionine is an important amino acid that acts as the initiator of protein synthesis (as N-formyl methionine) and of protein elongation. It is also the precursor of spermidine, a polyamine involved in cellular metabolism.[2] The met repressor is a dimer of identical 104 amino acid subunits, and is capable of repressing or depressing target genes within 30 minutes of a change of methionine concentration.[3]
Key structural features of the met repressorKey structural features of the met repressor
The met operators in E. coli consist of tandem repeats of eight base pair sequences, AGACGTCT, known as 'met boxes'. [2] These met boxes vary in length from 16-40 base pairs, corresponding to two to five met boxes. The consensus sequence is highly symmetrical, with centers of inverted repeats at the center of each met box and at the junctions between them. The , that the met repressor binds to in the E. coli met repressor-operator complex at the top of the page, has the sequence 5'-TTAGACGTCTAGACGTCTA-3' which contains two tandem consensus met boxes flanked by TA base pairs on the 3' end and one unpaired T on the 5' end.[4] One of the products of the methionine biosynthetic pathway is and it acts as a corepressor. The repressor has to bind two molecules of SAM non-cooperatively in order for the repressor to bind DNA. The following image represents SAM:
How met repressor interacts with DNAHow met repressor interacts with DNA
|
The E. coli methionine repressor was the first characterized member of the ribbon-helix-helix (RHH) class of DNA-binding proteins that uses a pair of beta-strands to interact with DNA bases. The [5] form a two-stranded anti-parallel beta sheet that inserts into the DNA's major groove. [1] The beta strands make sequence specific contacts with the DNA through hydrogen bonding.
There are three principle regions of the protein where there is direct contact with DNA. The first region includes the only direct contacts to the bases which are made by side chains of the beta strands lying in the major groove. hydrogen bind with DNA. The hydroxyl groups of Thr 25 and 25' donate hydrogen bonds to N7 of A3 and A11', respectively. [4] The Lys 23 hydrogen binds to O6 and N7 of G10'. The Lys 23' differs and only binds to O6 of G2. Both lysines also make water-mediated hydrogen bonds to adjacent base pairs, to N7 of A9' and O4 of T14', respectively. The van der Waals surface of the beta-strands makes a loose fit to the major groove. This leaves room for a number of solvent molecules to fit.
The second major contact region is the N terminus of the B helix. The N terminus interacts strongly with the 5' phosphate of G10', and the N terminus of the other B helix interacts strongly with the 5' phosphate of G2. [4] Hydrogen bonds form from the main-chain amides of of the two B helixes, and the side chain hydroxyl groups of the respective phosphates.
The third major region of direct interaction is the . [4] The main-chain amides of Gly 15 and Lys 17 of one B helix interact directly with the phosphate of T8', and the main-chain amides of Gly 15 and Lys 17 of the other B helix interact directly with the phosphate of T0.
The total repressor-operator complex has 8 direct hydrogen bonds from the two dimers to base pairs and 38 hydrogen bonds to phosphates. [4]
About this StructureAbout this Structure
1CMA is a 4 chains structure of sequences from Escherichia coli. Full crystallographic information is available from OCA.
ReferencesReferences
- ↑ 1.0 1.1 Voet, Donald, Judith G. Voet, and Charlotte W. Pratt. Fundamentals of Biochemistry Life at the Molecular Level. New York: John Wiley & Sons, 2008. p. 578-579. Print.
- ↑ 2.0 2.1 Phillips SE, Stockley PG. Structure and function of Escherichia coli met repressor: similarities and contrasts with trp repressor. Philos Trans R Soc Lond B Biol Sci. 1996 Apr 29;351(1339):527-35. PMID:8735275 doi:10.1098/rstb.1996.0051
- ↑ Augustus AM, Reardon PN, Spicer LD. MetJ repressor interactions with DNA probed by in-cell NMR. Proc Natl Acad Sci U S A. 2009 Mar 31;106(13):5065-9. Epub 2009 Mar 16. PMID:19289840 doi:10.1073/pnas.0811130106
- ↑ 4.0 4.1 4.2 4.3 4.4 Somers WS, Phillips SE. Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands. Nature. 1992 Oct 1;359(6394):387-93. PMID:1406951 doi:http://dx.doi.org/10.1038/359387a0
- ↑ Marincs F, Manfield IW, Stead JA, McDowall KJ, Stockley PG. Transcript analysis reveals an extended regulon and the importance of protein-protein co-operativity for the Escherichia coli methionine repressor. Biochem J. 2006 Jun 1;396(2):227-34. PMID:16515535 doi:10.1042/BJ20060021