Mediator
MediatorThe mediator or mediator of RNA polymerase II transcription subunit, found in the yeast S. cerevisiae, connects repressors and activators bound to regulatory DNA with RNA polymerase II (Pol II). As a result, the mediator is an important regulator of eukaryotic transcription.[1] Also, mediator is currently being studied because of its ability and role to regulate gene expression.[2] Mediator is in the form of three main modules, which are the tail, arm, and head. The of the mediator is split into three domains that go through significant conformational changes, and it interacts with the Rpb7 and Rpb4 subunits of Pol II and TATA binding protein subunit of transcription factor TFIID.[3]
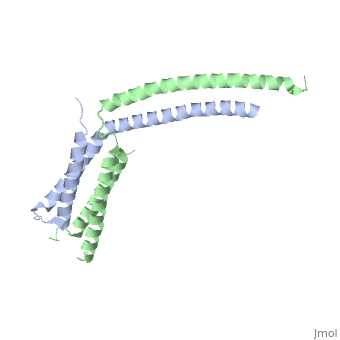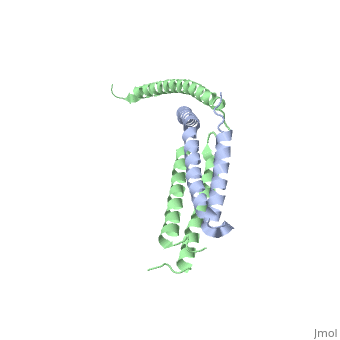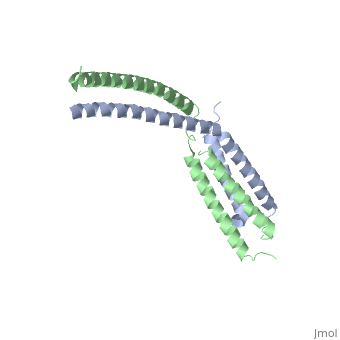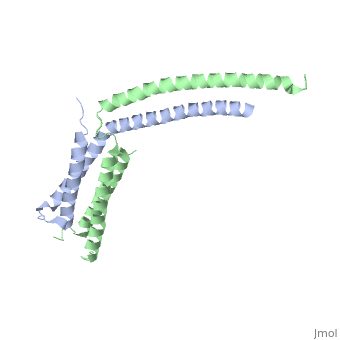
StructureThe of mediator from S.cerevisiae is made of seven subunits which are Med22(Srb6), Med20(Srb2), Med18(Srb5), Med17(Srb4), Med11, Med6, and Med8.[3] Of these subunits, Med11, Med17(Srb4), and Med22(Srb6) make the mini-head of the head module. The subunits of the core-head plus the subunits of the mini-head are what make up full head structure. The structure shows the three domains: the neck, which is shown by pink helices, the fixed jaw, which is shown by mostly pink helices and a few yellow beta sheets, and the movable jaw, which is shown by mostly yellow beta sheets and a few pink helices. The tail module of yeast mediator is made up of Med2, Med3, Med14, Med15 and Med16. The middle module of yeast mediator is made up of Med1, Med4, Med5, Med7, Med10, Med12, Med13, Med19, Med21, Med26, Med31, CycC and Cdk8.
|
| ||||||||||
3D structures of mediator3D structures of mediator
Updated on 27-June-2023
7emf, 7enj – hMed in mediator complex – human – Cryo EM
7ena, 7enc – hMed in TFIID-mediator complex – Cryo EM
7lbm, 7nvr – hMed in transcription pre-initiation complex – Cryo EM
2l23, 2l6u, 2ky6, 2xnf – hMed25 activator-interacting domain - NMR
5odd – hMed26 N terminal
6h02 – hMed23 + nanobody
6w1s – mMed in mediator complex – mouse – Cryo EM
3rj1 – yMed6 + Med8 + Med11 + Med17 + Med18 + Med20 + Med22 - yeast
7kpw – yMed12 – Cryo EM
7kpv, 7kpx – yMed12 + Med13 + SSN3 + suppressor of RNA polymerase – Cryo EM
4v1o – yMed8 in pre-initiation complex – Cryo EM
1yke, 1ykh – yMed7 + Med21
3fbi, 3fbn – yMed7 N terminal + Med31
2hzs – yMed8 C terminal + Med18 + Med20
3r84 - yMed11 + Med22 (mutant)
6aly – yMed 15 ABD2 residues 277-368
2lpb - yMed15 + general control protein GCN4
2hzm – yMed18 + Med20
5oqm – yMed4 + FTIIH in transcription pre-initiation complex – Cryo EM
5sva, 5u0s – fyMed in transcription pre-initiation complex – fission yeast – Cryo EM
5n9j, 5u0p – fyMed in mediator complex – Cryo EM
3c0t - fyMed8 C terminal + Med18
6xp5 – CtMed in head-middle complex – Chaetomium thermophilum – Cryo EM
7jmn – CtMed in tail complex – Cryo EM
ReferencesReferences
- ↑ Imasaki T, Calero G, Cai G, Tsai KL, Yamada K, Cardelli F, Erdjument-Bromage H, Tempst P, Berger I, Kornberg GL, Asturias FJ, Kornberg RD, Takagi Y. Architecture of the Mediator head module. Nature. 2011 Jul 3;475(7355):240-3. doi: 10.1038/nature10162. PMID:21725323 doi:10.1038/nature10162
- ↑ Taatjes DJ. The human Mediator complex: a versatile, genome-wide regulator of transcription. Trends Biochem Sci. 2010 Jun;35(6):315-22. Epub 2010 Mar 17. PMID:20299225 doi:10.1016/j.tibs.2010.02.004
- ↑ 3.0 3.1 Imasaki T, Calero G, Cai G, Tsai KL, Yamada K, Cardelli F, Erdjument-Bromage H, Tempst P, Berger I, Kornberg GL, Asturias FJ, Kornberg RD, Takagi Y. Architecture of the Mediator head module. Nature. 2011 Jul 3;475(7355):240-3. doi: 10.1038/nature10162. PMID:21725323 doi:10.1038/nature10162