Brr2
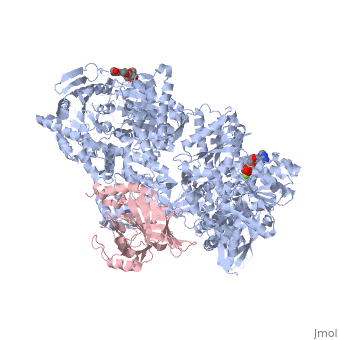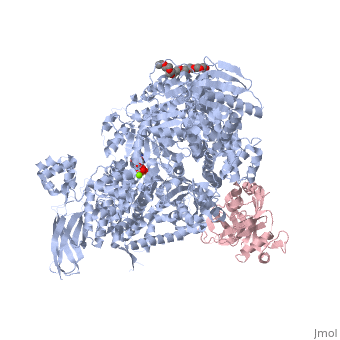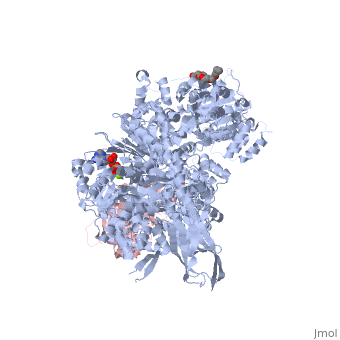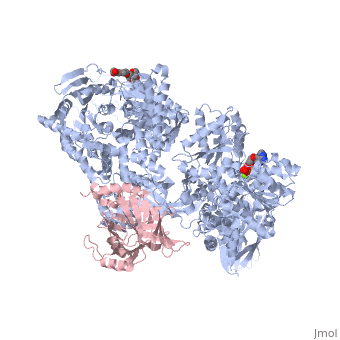
Structure of Brr2by Kelly Hrywkiw IntroductionThe processing of pre-mRNA takes place through the use of a large dynamic machine known as the spliceosome, through which introns are removed and exons are spliced together to create a mature mRNA[1] [2]. The spliceosome is comprised of five snRNA] molecules (snRNAs U1, U2, U4, U5, and U6) and over one hundred associated proteins[1][2]. Assembly of the spliceosome is thought to take place in a stepwise manner around the pre-mRNA transcript[1][2]. The first step involves recognition of the 5’ splice site by U1 snRNP, followed by recognition of the branch point sequence by U2 snRNP[1][2]. From this point the remaining snRNPs U4, U5, and U6 join as a preformed tri-snRNP[1][2]. Together the five snRNPs form the precatalytic spliceosome which must undergo a series of changes before it can actively splice[3][2][4][1]. A family of proteins that plays a large role in activating the spliceosme is the helicase superfamily 2 (SF2)[4][3]. Within this family over ten DExD/H-box proteins are associated with various stages of conformational rearrangement necessary for spliceosome activation[4]. These proteins are thought to destabilizing short RNA duplexes in a nonprocessive manner, or alter the RNA protein interactions of the various snRNPs[4]. In this rearrangement process the U4/U6 di-snRNP contains a long duplex that needs to be unwound, however most DExD box proteins can only unwind duplex with less than two turns[4]. The unwinding of U4/U6 takes place by Brr2, a DExD/H-box protein associated with U5 snRNP, which is unlike other spliceosomal RNA helicases as it belongs to the Ski2 subfamily of the helicase SF2[4][3]. Structure of Brr2 Brr2 contains an N-terminal domain which is thought to have little tertiary structure (Fig 1)[4][3]. In addition, is contains two helicase cassettes (residues 496-1310 and 1311-2162) which is outside the norm, as all but one other known DExD/H-box protein contain but one helicase cassette (Fig 2)[3]. Each cassette contains dual RecA-like domains and a Sec63 domain (Fig 2)[4]. While RecA domains are common to all helicase SF2 proteins Sec63 domain which exhibits a similar sequence to the Sec63 protein, that is essential in the protein-translocation apparatus in the endoplasmic reticulum, are not a common[4][3]. The RecA-like domain is connected to the Sec63 domain via a WH connector[4]. The N-terminal cassette is critical for ATPase activity and U4/U6 unwinding. The C-terminal cassette can undergo catalytically harmful mutations without drastically impairing the overall function of Brr2, and is thought to be involved in protein-protein interactions[4][3]. The Sec63 DomainTo date the only crystal structures of Brr2 are that of the domain in the C-terminal helicase cassette (Sec63c) which contains three sections. The (residues 1859-1990) is comprised of six α helices and one 310 helix. The longest of the helices is α5 which gives stability to the other helices, such that they are able to form a helical bundle through a series of hydrophobic contacts. The (residues 1991-2048) exhibits a helix loop helix fold comprised of four α helices and one 310 helix. The (residues 2049-2163) resembles a seven-stranded immunoglobulin-like β sandwich. Before the N-terminal domain is a region of residues which is highly flexible and differs between different crystalized versions of Sec63c, however other than this region there is high similarity between the crystal structures. All three domains are in contact with one another. The primary element that appears the fix the domains together is the β sandiwich, specifically the loops connecting β2 and β3, and β6 and β7 which are located towards the center of Sec63c[4].
Structural Similarity to Hel308Hel308 is part of the SF2 helicase protein superfamily that plays a role in DNA repair, genome stability and recombination[5]. Interestingly, there is a significant amount of structural homology between the N-terminal and central domains of Sec63c and domains and of Hel308[4][3]. In addition Hel308 contains two RecA domains with sequence similarity to those in Brr2[4][3]. To reiterate, Hel308 has five domains, where domains 1-3 are closely related to the two RecA-like domains and the WH connector, and domain 4 and 5 are similar to the N-terminal and central domains of Sec63c. One difference of note is that Hel308 does not contain the β sandwich in the Sec63c domain. This suggests that the Brr2 is composed of two Hel308-like modules, where each module is analogous to a cassette and an N-terminal domain (Fig.1)[4][3]. The sequence similarity and proposed strucutre between the N-terminal cassette and Hel308 is greater than that between Hel308 and the N-terminal cassette[3]. Using the structural knowledge of Hel308 in tandem with Brr2 it is possible to form a better understanding of how Brr2 functions. Unwinding the U4/U6 duplexThe first four domains of Hel308 form a ring containing a which can pass through[4][3][6]. In this pore the central helix contains that face the interior of the pore and are predicted to interact with DNA[4]. The proposed mechanism by which Hel308 unwinds DNA duplexes is as follows: through the binding of ATP the fourth motif in domain 2 pushes the DNA in the direction of domain 1 while domain 2 also pushes on the region of the DNA, and the changes in domains 2 and 4 may be enough movement to slip the 3’ region across motif 1a[5]. ATP hydrolysis would bring domain two back to its original conformation to await another ATP[5]. Furthermore, the β hairpin located between domains 4 and five is thought to disrupt 2bp of the DNA duplex[4][3]. Overall, this suggests that Hel308 unwinds DNA through a processive manner[4][3]. While the exact sequence and structure of the N-terminal cassette is not a perfect match to Hel308 it is more similar than the C-terminal cassette and therefore may exhibit a similar processive unwinding mechanism that would be important in unwinding the long U4/U6 RNA duplex. The C-terminal cassette has little to no helicase and ATPase activity, however has been shown to interact with other splicing factors such as Prp8 and Snu114[3]. This suggest that it may play an important role in protein-protein interactions. |
| ||||||||||
Additional ResourcesAdditional Resources
- Crystal structure of the second Sec63 domain of yeast Brr2, in the RCSB Protein Data Bank
- Structure of the C-terminal Sec63 unit of yeast Brr2, P41212 Form, in the RCSB Protein Data Bank
- Structure of the C-terminal Sec63 unit of yeast Brr2, P212121 Form, in the RCSB Protein Data Bank
- DNA REPAIR HELICASE HEL308,in the RCSB Protein Data Bank, in the RCSB Protein Data Bank
- Apo structure of the Hel308 superfamily 2 helicase, in the RCSB Protein Data Bank
- Crystal structure of superfamily 2 helicase Hel308 in complex with unwound DNA, in the RCSB Protein Data Bank
3D structures of Brr23D structures of Brr2
See Helicase
ReferencesReferences
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 van der Feltz C, Anthony K, Brilot A, Pomeranz Krummel DA. Architecture of the Spliceosome. Biochemistry. 2012 Apr 10. PMID:22471593 doi:10.1021/bi201215r
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 Sperling J, Azubel M, Sperling R. Structure and function of the Pre-mRNA splicing machine. Structure. 2008 Nov 12;16(11):1605-15. PMID:19000813 doi:10.1016/j.str.2008.08.011
- ↑ 3.00 3.01 3.02 3.03 3.04 3.05 3.06 3.07 3.08 3.09 3.10 3.11 3.12 3.13 3.14 Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, Guthrie C, Pleiss JA, Zhao R. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol. 2009 Jul;16(7):731-9. Epub 2009 Jun 14. PMID:19525970 doi:10.1038/nsmb.1625
- ↑ 4.00 4.01 4.02 4.03 4.04 4.05 4.06 4.07 4.08 4.09 4.10 4.11 4.12 4.13 4.14 4.15 4.16 4.17 4.18 Zhang L, Xu T, Maeder C, Bud LO, Shanks J, Nix J, Guthrie C, Pleiss JA, Zhao R. Structural evidence for consecutive Hel308-like modules in the spliceosomal ATPase Brr2. Nat Struct Mol Biol. 2009 Jul;16(7):731-9. Epub 2009 Jun 14. PMID:19525970 doi:10.1038/nsmb.1625
- ↑ 5.0 5.1 5.2 Buttner K, Nehring S, Hopfner KP. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat Struct Mol Biol. 2007 Jul;14(7):647-52. Epub 2007 Jun 10. PMID:17558417 doi:10.1038/nsmb1246
- ↑ Richards JD, Johnson KA, Liu H, McRobbie AM, McMahon S, Oke M, Carter L, Naismith JH, White MF. Structure of the DNA repair helicase hel308 reveals DNA binding and autoinhibitory domains. J Biol Chem. 2008 Feb 22;283(8):5118-26. Epub 2007 Dec 4. PMID:18056710 doi:10.1074/jbc.M707548200