User:James D Watson/Structural Templates
Motifs In ProteinsMotifs In Proteins
The term "motif" when used in structural biology tends to refer to one of two cases:
- A particular amino-acid sequence that characterises a biochemical function
- A set of secondary structure elements that defines a functional or structural role
There are a great number of protein sequence motifs identified, many of which have well defined structural or functional roles. One such example of this is the so-called zinc finger motif which is readily identified from the following consensus sequence pattern (where "X" represents any amino acid):
Cys - X(2-4) - Cys - X(3) - Phe - X(5) - Leu - X(2) - His - X(3) - His
|
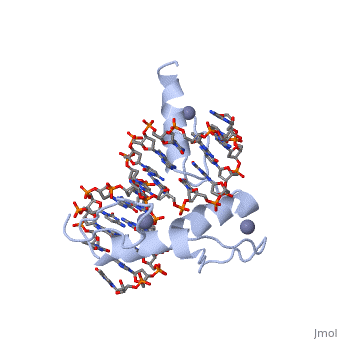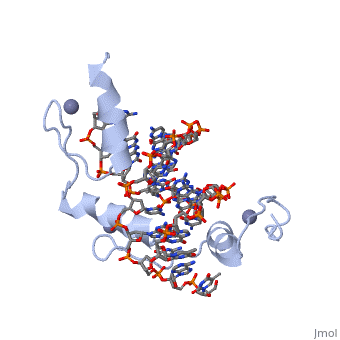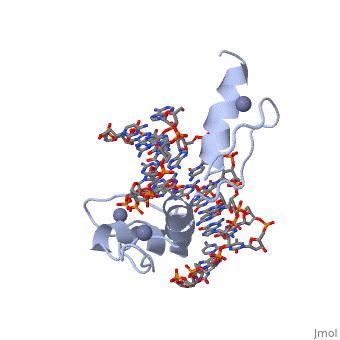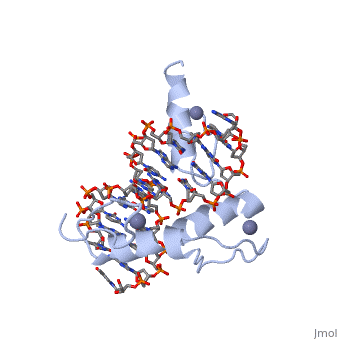
The example structure shown to is that of Zif268 protein-DNA complex from Mus musculus (PDB entry 1AAY). In this example (a C2H2 class zinc finger) the conserved and residues form ligands to a whose coordination is essential to stabilise the tertiary fold of the protein. The fold is important because it helps orientate the to bind to the .
However, there are also a number of repeated patterns and functional motifs revealed when the protein structure is examined. This page aims to introduce some of the main types of motif illustrating them on protein structures from the PDB.
Secondary structure elementsSecondary structure elements
Proteins are formed from linear chains of amino acids joined together by peptide bonds. These chains then fold up to form the three dimensional shape. However, the relative rigidity of the peptide bond combined with the presence of amino acid sidechains, means that not all conformations are acceptable and there are many cases where the various atoms in the chain start to collide with one another. Two of the most stable (and therefore most commonly observed) conformations are the α-helix and the β-pleated sheet. These are known as secondary structures.
α-helices The α-helix is formed when the amino acid backbone forms a right handed spiral with 3.6 amino acids per turn. The sidechains point outward, away from the centre of the helix, where athey can interact with solvent, other protein, small molecules or macromolecules. The structure is stabilised by regular hydrogen bonds that form between the backbone carbonyl oxygens and amide hydrogens. The bonding pattern for the α-helix is characterised by the carbonyl group of residue i hydrogen interacting with the amide group of residue i+4, this is known as an (i, i+4) interaction. The alpha-helix can take other less common forms including π-helices, 310-helices and their left handed forms (see table 1 for the helix parameters)
β-sheets occur in two varieties: parallel and anti-parallel. Different kinds of turns in the chain are also classified as secondary structures. A fourth and final type of secondary structure, known as random coil, refers to folds of the protein that do not fit into a classification. About 50% of all protein structure comes under this category.
Peptide Bonds Peptide bonds are formed as a protein is synthesized, between the carboxylic acid group of one amino acid and the amimo group of the next. Peptide bonds are rigid, planar bonds due to a degree of resonance that gives them a partial double bond character. There is no rotation around a peptide bond, so it confers some limitations to the folding of a protein chain.
β-sheets
β sheets are compact and stable structures. They are formed when two or more lengths of a protein chain lie next to each other so as to form hydrogen bonds between their respective backbones. In order for the backbones to be close enough for hydrogen bonds to form, the sidechains must not come between the backbones. Each length that participates in a β sheet is called a β strand.
There are two ways the strands can orient themselves to form &beta sheets: parallel and anti-parallel.
Anti-parallel β Sheets
Protein chains are synthesized starting at the amino terminus and ending at the carboxyl terminus. Thus a protein chain has a directionality. In an anti-parallel β sheet, the beta strands are aligned next to each other, running in opposite directions.
Hydrogen bonding
The backbone hydrogen bonding partners are the carbonyl oxygens and amide hydrogens. They are lined up nearly directly across from each other when the strands of a sheet are anti-parallel. They form fairly straight hydrogen bonds.
Sidechain conformation
The sidechains project out of the plane of the sheet, each consecutive sidechain emerging from the opposite side of the sheet.
Parallel β sheets
When β strands are in the parallel orientation, i.e., they run alongside each other in the same direction, the carbonyl oxygen and the amide protons are staggered, and the hydrogen bonds are on an angle across to the opposite strand.
As with the α-helix, the sidechains of the β sheet are free to interact with the environment, and the tertiary structure of the protein will be influenced by the sidechains projecting out of the β-sheet.
Secondary structure and the Jmol Menu
The secondary structure of a protein can be quickly seen using menu commands. First click View Animation to reset the display.
Select | All
Color | Cartoon | By Scheme | Secondary Structure
The α-helices are magenta, β sheets are yellow, and random coil is white.
QUESTIONS
The following interactive question(s) require you to interact with the structure to arrive at the correct answer. You may use any of the visualization controls or the dropdown menus to help you to answer the questions - direct manipulation of the structure may be required.
Question 1- Load structure
The α heilix and β sheets we've been looking at are parts of the ribosomal protein L9. It is composed of two globular domains with a very long α-helix between them. Given this image of L9 in spacefill, colored by element, use the Jmol menu to change the display to so that you can clearly see both (1) the pattern of the protein chain and (2) the default colors for secondary structure.
View Answer
Question 2 - Load structure Explore the Jmol menu to find commands relating to hydrogen bonds. Given this display of the backbone of ribosomal L9, display the hydrogen bonds that stabilize secondary structres. View Answer
|
|