User:James D Watson/Structural Templates
'Bold text'==Structural Motifs In Proteins== There are a great number of protein sequence motifs which have been found, many of which have a functional role. However, there are also a number of repeated patterns and functional motifs revealed when the protein structure is examined. This page aims to introduce various types of motif and illustrate them using the green links in the text.
Global foldGlobal fold
| |||||||||
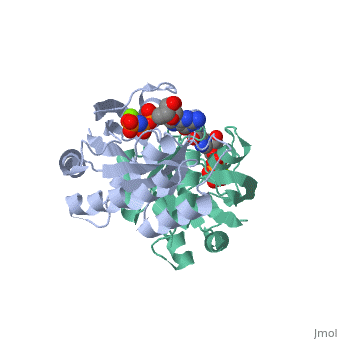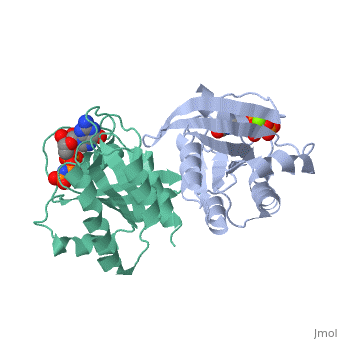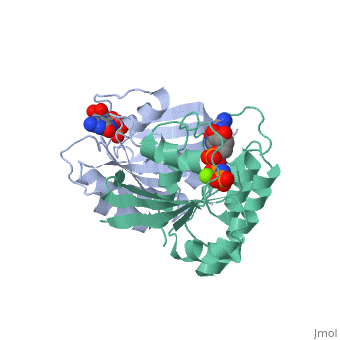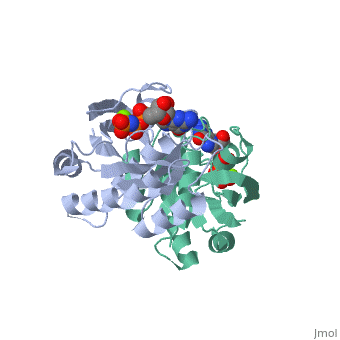
| 5p21, resolution 1.35Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
The image to the right shows the crystal structure of the H-ras oncogene protein p21 complexed to a substrate analogue (GppNp).
About this StructureAbout this Structure
5P21 is a Single protein structure of sequence from Homo sapiens. Full crystallographic information is available from OCA.
ReferenceReference
Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 A resolution: implications for the mechanism of GTP hydrolysis., Pai EF, Krengel U, Petsko GA, Goody RS, Kabsch W, Wittinghofer A, EMBO J. 1990 Aug;9(8):2351-9.
Structural superpositionStructural superposition
The viewer below left shows the structural superposition of the triphosphate conformation of H-ras p21 (PDB entry 5p21 - coloured orange) with Gdp-bound human rab21 gtpase (PDB entry 1z0i - coloured blue), the structural superposition was made using the MSDfold(SSM)1 server at the EBI2. Note that the global fold of these two proteins is almost identical yet their sequence identity is only 29.6% (as determined using FASTA). The viewer below right shows the p-loops of both structures superposed. The ligands bound superpose particularly well and comparison of the two p-loops loops show significant structural similarity but also highlights the sequence differences between the two proteins.
|
|
This is a placeholderThis is a placeholder
This is a placeholder text to help you get started in placing a Jmol applet on your page. At any time, click "Show Preview" at the bottom of this page to see how it goes.
Replace the PDB id after the STRUCTURE_ and after PDB= to load and display another structure.
| |||||||||
| 3cin, resolution 1.70Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Gene: | TM1419, TM_1419 (Thermotoga maritima MSB8) | ||||||||
| Activity: | Inositol-3-phosphate synthase, with EC number 5.5.1.4 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||