Prion protein
The prion protein (PrP) is a cell surface glycoprotein, which can exist in two alternatively folded confirmations: the cellular isoform (PrPC) can undergo a structural conversion to a 'scrapie' or disease associated isoform termed PrPSc.
Prion diseasesPrion diseases
The naturally ocuring prion diseases include Creutzfeldt Jakob disease (CJD) in people, bovine spongiform encephalopathy (BSE) commonly known as "mad cow" disease,scpie in sheep and goats, and chronic wasting disease in cervids are characterterized by aggregates of PrPSc. The spontaneous, genetic and infectious etiologies of prion diseases can be explained by a simple protein-based model in which PrPC is converted into PrPSc, which then initiates autocatalytic refolding of PrPC in a template-dependent manner.
In sporadic disease, the spontaneous refolding or misfolding of PrPC into PrPSc initiates the cascade. In genetic prion diseases, point mutations in PrP make this more likely to happen than in the wild type protein, Infectious etiology is explained by introduction of exogenous PrPSc which then initiated refolding of endogenous PrPC.
Structure of PrPCStructure of PrPC
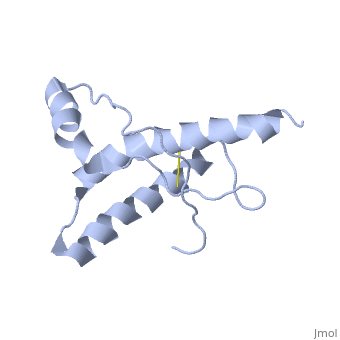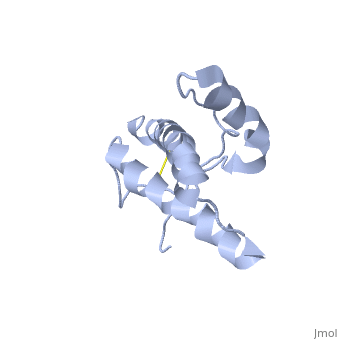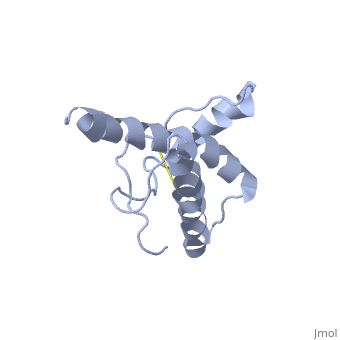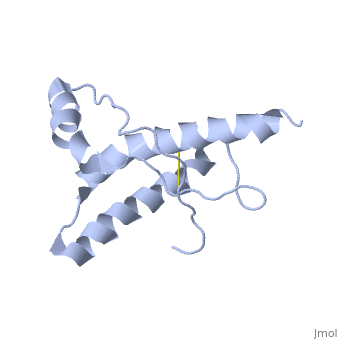
| |||||||||
| 1hjm, 1 NMR models () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Related: | 1e1g, 1e1j, 1e1p, 1e1s, 1e1u, 1e1w, 1hjn | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
PrPC has a natively unstructured N-terminal region, and a predominantly α-helical C-terminal region from residues ~120-230, with a single disulfide bond. The presence of the N-terminal region has little impact on the structure of the C-terminal domain [1].
The structure is highly conserved amongst mammals, and only differs slightly in birds, reptiles and amphibians. The vast majority of structures have been determined by
Although having a similar overall fold, the X-ray structure of sheep PrP was dimeric
Models of PrPSc structureModels of PrPSc structure
Circular dichroism studies first demonstrated that PrPSc had very different proportions of α-helices and β-sheet to PrPC
There are a number of technical obstacles in determining the molecular structure of PrPSc
Genetic prion diseasesGenetic prion diseases
A number of mutations in PrP have been identified which correlate with a high incidence of prion disease. The structure of HuPrP,E200K was determined nd shown to be To date, structural studies of all mutant PrPC have extremely similar structures to wild type PrPC, suggesting a kinetic basis for the difference in converting to PrPSc.
Prion strainsPrion strains
The strain phenomenon of prions ( ) was initially difficult to equate with the
Selected PrP structuresSelected PrP structures
All structures determined by NMR unless otherwise specified
Human PrPHuman PrP
- 1qlx HuPrP residues 23-230
- 1qm0 HuPrP residues 90-230
- 1qm2 HuPrP residues 121-230
- 1i4m HuPrP residues 119-226 (determined by X-ray crystallography)
- 1fkc HuPrP,E200K residues 90-231 (genetic prion disease)
- 1h0l HuPrP residues 121-230, with an additional disulphide bond analogous to the homolog Doppel