User:Adrian Aldrich/Prokaryotic Glutamine Synthetase Pfam Domains Outline
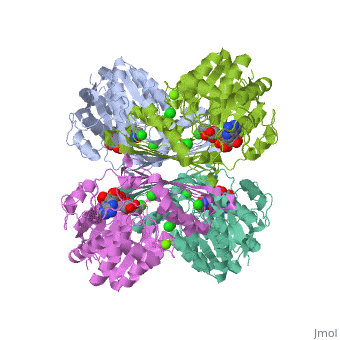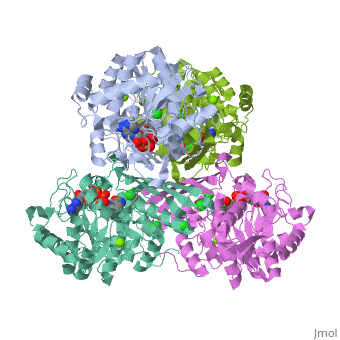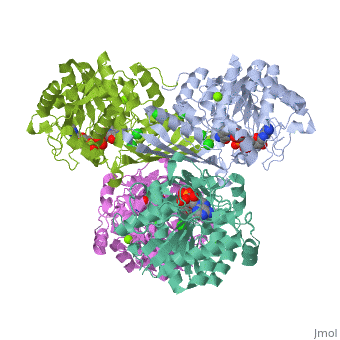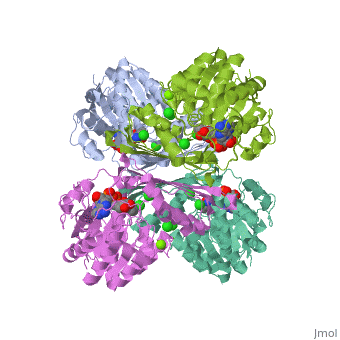
Prokaryotic Glutamine Synthetase Tertiary Structure: PFam domains
-(describe structure of protein leading in to a single chain) -each chain contains 2 domains, the Glutamine synthetase beta grasp domain, and the glutamine synthetase catalytic domain. -the two domains are preceeded by a meander of 3-24 residues that link the chains in the proteins core.
Beta Grasp domain -comprised of residues 43-123(verification needed for using structure viewer) -binds glutamate, ammonia, and ATP
<show protein alignment image> <show structure of the domain in viewer>
Catalytic domain -comprised of residues 129-382 -how it functions is not described, more research is necessary here
<show alignment image> <show structure of the domain in viewer>
Active site -there are 10 active sites in the decamer of the protein, each formed from the beta grasp domain of one chain and the catalytic domain of the neighboring chain.
<show relation between chains, domains, and active sites with viewer> -describe visually the important aspects of the active site <show active site and ligands relationship, may need multiple scenes>
Replace the PDB id after the STRUCTURE_ and after PDB= to load and display another structure.
| |||||||||
| 3cin, resolution 1.70Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , | ||||||||
| Gene: | TM1419, TM_1419 (Thermotoga maritima MSB8) | ||||||||
| Activity: | Inositol-3-phosphate synthase, with EC number 5.5.1.4 | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum, TOPSAN | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||