2x0n
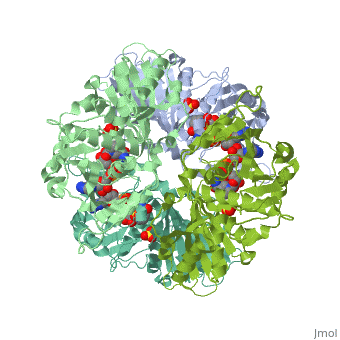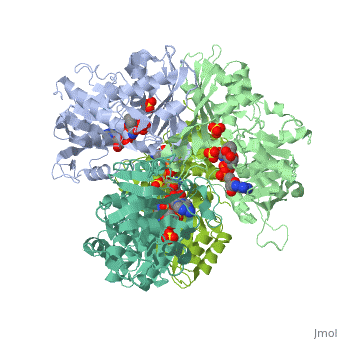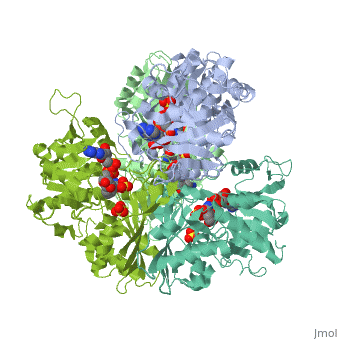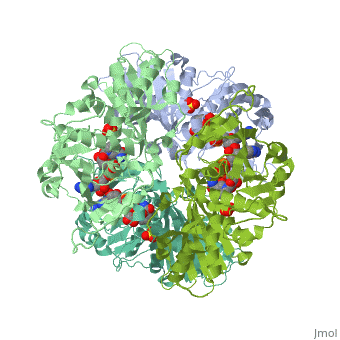
Structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei determined from Laue dataStructure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei determined from Laue data
Structural highlights
FunctionEvolutionary ConservationCheck, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedThe three-dimensional structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase [D-glyceraldehyde-3-phosphate:NAD+ oxidoreductase (phosphorylating), EC 1.12.1.12] from the sleeping-sickness parasite Trypanosoma brucei was solved by molecular replacement at 3.2-A resolution with an x-ray data set collected by the Laue method. For data collection, three crystals were exposed to the polychromatic synchrotron x-ray beam for a total of 20.5 sec. The structure was solved by using the Bacillus stearothermophilus enzyme model [Skarzynski, T., Moody, P. C. E. & Wonacott, A. J. (1987) J. Mol. Biol. 193, 171-187] with a partial data set which was 37% complete. The crystals contain six subunits per asymmetric unit, which allowed us to overcome the absence of > 60% of the reflections by 6-fold density averaging. After molecular dynamics refinement, the current molecular model has an R factor of 17.6%. Comparing the structure of the trypanosome enzyme with that of the homologous human muscle enzyme, which was determined at 2.4-A resolution, reveals important structural differences in the NAD binding region. These are of great interest for the design of specific inhibitors of the parasite enzyme. Structure of glycosomal glyceraldehyde-3-phosphate dehydrogenase from Trypanosoma brucei determined from Laue data.,Vellieux FM, Hajdu J, Verlinde CL, Groendijk H, Read RJ, Greenhough TJ, Campbell JW, Kalk KH, Littlechild JA, Watson HC, et al. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2355-9. PMID:8460146[1] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||