2dpd
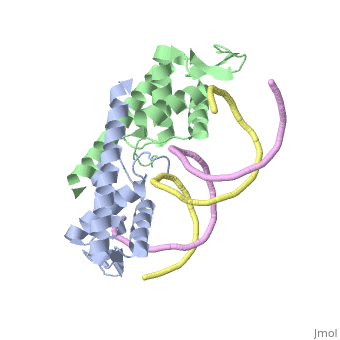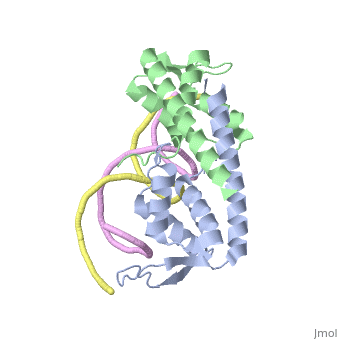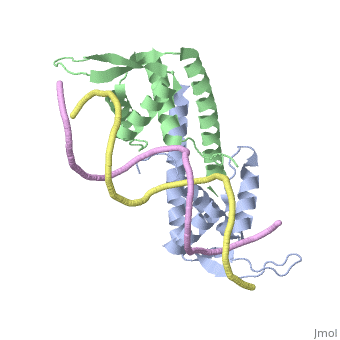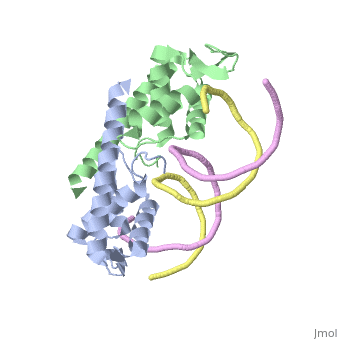
Crystal structure of the Replication Termination Protein in complex with a pseudosymmetric B-siteCrystal structure of the Replication Termination Protein in complex with a pseudosymmetric B-site
Structural highlights
FunctionRTP_BACSU Plays a role in DNA replication and termination (fork arrest mechanism). Two dimers of rtp bind to the two inverted repeat regions (IRI and IRII) present in the termination site. The binding of each dimer is centered on an 8 bp direct repeat. See Also |
| ||||||||||||||||