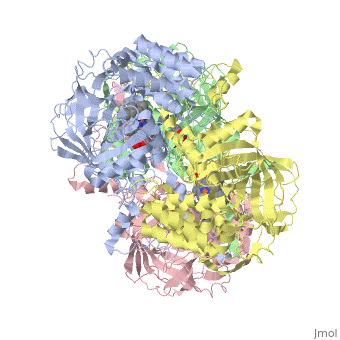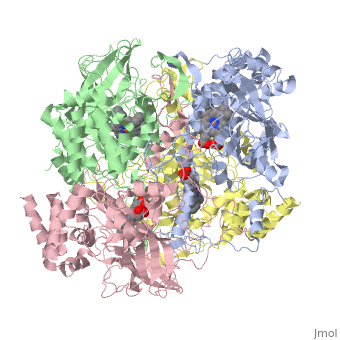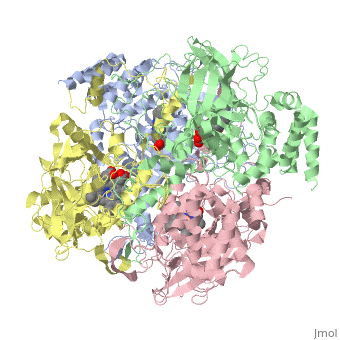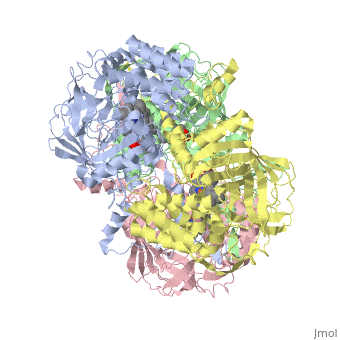1qqw
CRYSTAL STRUCTURE OF HUMAN ERYTHROCYTE CATALASECRYSTAL STRUCTURE OF HUMAN ERYTHROCYTE CATALASE
Structural highlights
Disease[CATA_HUMAN] Defects in CAT are the cause of acatalasemia (ACATLAS) [MIM:614097]. A metabolic disorder characterized by absence of catalase activity in red cells and is often associated with ulcerating oral lesions.[1] Function[CATA_HUMAN] Occurs in almost all aerobically respiring organisms and serves to protect cells from the toxic effects of hydrogen peroxide. Promotes growth of cells including T-cells, B-cells, myeloid leukemia cells, melanoma cells, mastocytoma cells and normal and transformed fibroblast cells.[2] Evolutionary ConservationCheck, as determined by ConSurfDB. You may read the explanation of the method and the full data available from ConSurf. Publication Abstract from PubMedCatalase (E.C. 1.11.1.6) was purified from human erythrocytes and crystallized in three different forms: orthorhombic, hexagonal and tetragonal. The structure of the orthorhombic crystal form of human erythrocyte catalase (HEC), with space group P2(1)2(1)2(1) and unit-cell parameters a = 84.9, b = 141.7, c = 232.5 A, was determined and refined with 2.75 A resolution data. Non-crystallographic symmetry restraints were employed and the resulting R value and R(free) were 0.206 and 0.272, respectively. The overall structure and arrangement of HEC molecules in the orthorhombic unit cell were very similar to those of bovine liver catalase (BLC). However, no NADPH was observed in the HEC crystal and a water was bound to the active-site residue His75. Conserved lattice interactions suggested a common growth mechanism for the orthorhombic crystals of HEC and BLC. Structure of human erythrocyte catalase.,Ko TP, Safo MK, Musayev FN, Di Salvo ML, Wang C, Wu SH, Abraham DJ Acta Crystallogr D Biol Crystallogr. 2000 Feb;56(Pt 2):241-5. PMID:10666617[3] From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine. See AlsoReferences
|
| ||||||||||||||||||||