Introduction to Jmol
What is Jmol?What is Jmol?
Jmol is a free, open-source program that allows the viewer to see chemical structures in three dimensions. Jmol previously used Java, but has now switched to HTML5-only.
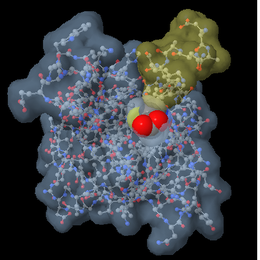
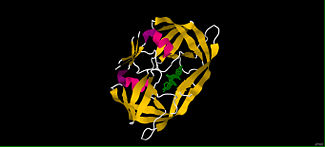
What makes this program very useful is Jmol's ability to manipulate the 3D models and display the molecules as "ball and stick," "space filling," "ribbon," "cartoon," etc. One can also do a combination of any or all possible displays. For example: one can display a ligand as "ball and stick" while displaying the protein its interacting with in a "cartoon" display.
In order to use Jmol you can start from scratch by building your own molecule or you can download one from the RCSB Protein Data Bank.
Who uses Jmol?Who uses Jmol?
Many people use Jmol as an educational, chemistry and biochemistry tool to help visualize molecules and compounds. Many websites, books and even blogs use Jmol as we can see from this site: [[1]]
Example of JmolAs an example we will be using the drug darunavir. Darunavir (Prezista) was approved in 2006 for the treatment of HIV-1. Darunavir is a second generation protease inhibitor. Proteases are used during the HIV replication in order to cut proteins the viruses need in order to survive. Protease inhibitors work by "plugging" the protease and not allowing the HIV proteins to be cut; thus, inhibiting HIV synthesis. This drug is a good example, because it will allow us to view how the drug is able to fit into a pocket withing the protease. The first scene that we see here is one pulled from RCSB Protein Data Bank. This scene demonstrates the secondary structure of the . The ligand and water molecules have been hidden in order to view the protein alone. This scene shows the in which darunavir fits into. After inverting the selection and targeting the ligand we can visualize .
|
| ||||||||||