Polygalacturonase
|
Polygalacturonases (PGs) catalyze the enzymatic depolymerization of pectins – polysaccharides that comprise the plant cell wall. Polymer disassembly of substrates by exo- and endo- PGs is carried out via a hydrolytic mechanism. Degradation of pectins in plant cell walls contributes to ripening of fruits, such as tomatoes and melons (Polygalacturonases: many genes in search of a function). Microbial PGs have been identified to be a part of defense mechanisms because of their role in pathogen attack (crystal structure).
FunctionFunction
Polygalacturonases hydrolyze α-(1-4) – glycosidic bonds between consecutive galacturonic acid residues in polygalacturonic acids. Structural variation has been identified among differing PGs depending on organismal origins and catalytic functions. For example, endo-polygalacturonases produced from Erwinia carotovora demonstrate functional similarity to pectate lyases in that they cleave polygalacturonic acids in a calcium-depended manner via β-elimination (crystal structure).
DiseaseDisease
RelevanceRelevance
Structural highlightsStructural highlights
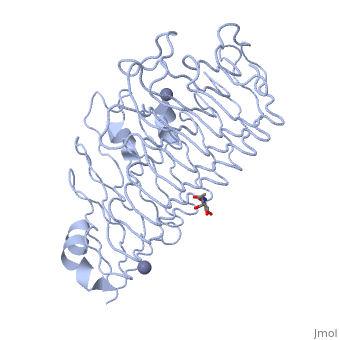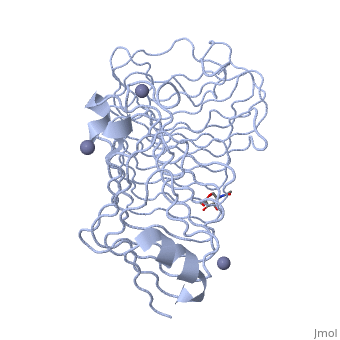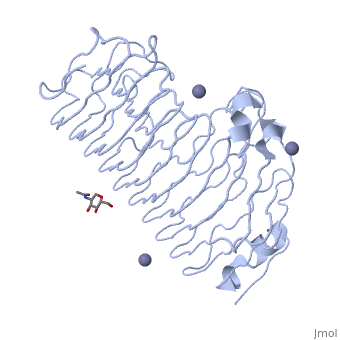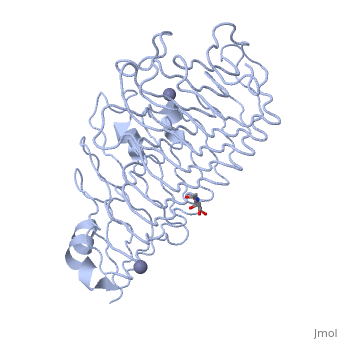
The secondary structure of PGs is comprised of a ten turn right handed beta helix domain along with two loop regions that together form the substrate-binding cleft, which appears to have a tunnel-like shape. The active site of PGs is found between the two looped regions of the protein. Located within the looped regions are two conserved aspartate residues that are predicted to participate in catalytic activity (crystal structure).