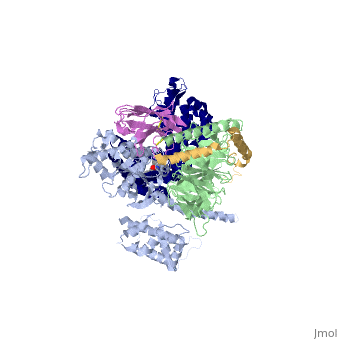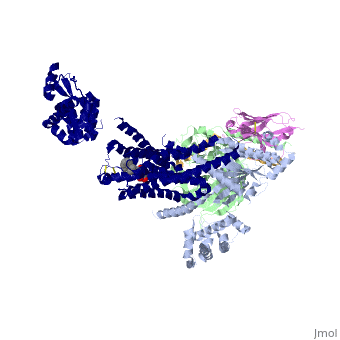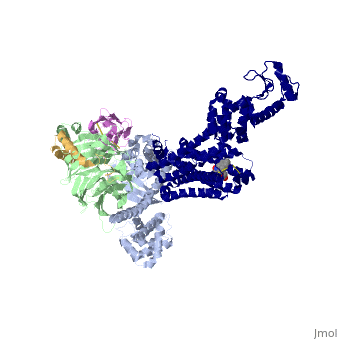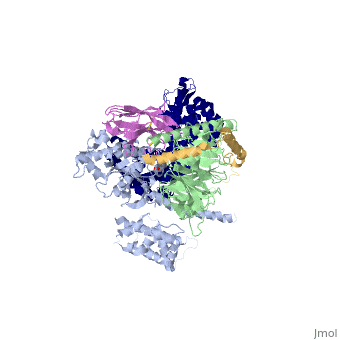
Beta2 adrenergic receptor-Gs protein complex
Beta2 adrenergic receptor-Gs protein complexBeta2 adrenergic receptor-Gs protein complex
IntrodcutionYou may include any references to papers as in: the use of JSmol in Proteopedia [1] or to the article describing Jmol [2] to the rescue. G protein-coupled receptors (GPCRs) are a large family of protein receptors which have seven-transmembrane helices and are found over a large array on eukaryotic cells. These receptors take major part in a multitude of signal transduction pathways, including amongst others responses to hormones and neurotransmitters, sensing light, taste and smell, and many other responses. These receptors are also involved in many different types of diseases and are the target of almost 50% of current medical drugs. The Beta-2 Adrenergic Receptors are a type of GPCRs which are activated by catecholamine hormone ligands such as adrenaline (epinephrine). These receptors are responsible for many of the adrenaline related (“fight-or-flight”) responses and functions, and are being a common model system for the GPCR family. GPCRs bind their ligand and overcome a conformational change which allows a Guanine nucleotide-binding protein (G protein) to detach from the cellular end of the receptor and start the different signal transduction pathways. Since these receptors have seven transmembrane helices as well as inner and outer cell regions, they are very difficult to purify and crystalize. Some crystal structures have been determined for the inactive receptors as well as for the G protein that they bind. 3sn6 is the first structure of the full complex of the Beta 2 Adrenergic Receptor bound to Gs in their active state, and it provides the first high-resolution insight into the mechanism of signal transduction across the plasma membrane by a GPCR. Obtaining the structure3sn6 is a 5 chain structure sequenced from Bos taurus, Enterobacteria phage t4, Lama glama and Rattus norvegicus. The b2AR and Gs couple efficiently in lipid bilayers, but not in detergents used to solubilize and purify these proteins. In order to overcome that, different approaches were used, including replacement of the unstructured amino terminus of the b2AR with T4 lysozyme (T4L). Another problem was that the two domains that make the G alpha s subunit and form the nucleotide- binding pocket of the G alpha subunit have great position variability when not bound to the guanine nucleotide, and since GTP and GDP disrupt the formation of the complex alternative stabilization methods where carried out, including obtaining the nanobody (a single domain antigen binding fragments obtained from Llamas) Nb35. The final T4L–b2AR–Gs–Nb35 complex was used to obtain crystals that grew to 250 mm in LCP and diffracted to 2.9A˚. A 3.2A˚ data set was obtained from 20 crystals and the structure was determined by molecular replacement Proteins. Structure of the active-state b2ARComparing this structure of the active state agonist-bound receptor to a structure of an inactive receptor reveals most notably a 14A˚ outward movement of TM6 when measured at the Calpha carbon of E268. There is a smaller outward movement and extension of the cytoplasmic end of the TM5 helix by 7 residues. A stretch of 26 amino acids in the third intracellular loop (ICL3) is disordered. Another notable difference between inactive and active structures is the second intracellular loop (ICL2), which forms an extended loop in the inactive b2AR structure and an alpha-helix in the b2AR–Gs complex. However, this helix may not be a feature that is unique to the active state, because it is also observed in the inactive structure of the highly homologous avian b1AR. Interface of the receptor and GsThe active state of the b2AR is stabilized by extensive interactions with the GasRas domain. There are no direct interactions with the Gbeta or Ggamma subunits. The total buried surface of the b2AR–GasRas interface is 2,576A˚ (1,300A˚ for GasRas and 1,276A˚ for the b2AR). This interface is formed by ICL2, TM5 and TM6 of the b2AR, and by a5-helix, the aN–b1 junction, the top of the b3-strand, and the a4-helix of GasRas. Some of the b2AR sequences involved in this interaction have been shown to have a role in G protein coupling; however, there is no clear consensus sequence for Gs-coupling specificity when these segments are aligned with other GPCRs. Structure of activated GsA surprising observation in the b2AR–Gs complex is the large displacement of theGasAH relative toGasRas (an approximately 127 degree rotation about the junction between the domains). The nucleotide-binding pocket is formed by the interface between GasRas and GasAH, and guanine nucleotide binding stabilizes the interaction between these two domains. The loss of this stabilizing effect of guanine nucleotide binding is consistent with the high flexibility observed for GasAH. Comparing the structure of GasRas from the b2AR–Gs complex with that from a Gas–GTPgammaS complex shows the largest difference in the a5-helix, which is displaced 6A˚ towards the receptor and rotated as the carboxy-terminal end projects into transmembrane core of the b2AR. Associated with movement of the a5-helix, the b6-a5 loop, which interacts with the guanine ring in the Gas–GTPcS structure, is displaced outward, away from the nucleotide-binding pocket. The movement of the a5-helix is also associated with changes in interactions between this helix and the b6-strand, the aN-b1 loop, and the a1-helix. The b1-strand forms another link between the b2AR and the nucleotide-binding pocket. There are further changes in the b1-a1 loop (P-loop) that coordinates the b-phosphate in the GDP and GTP-bound forms. See Also
|
| ||||||||||
ReferencesReferences
- ↑ Hanson, R. M., Prilusky, J., Renjian, Z., Nakane, T. and Sussman, J. L. (2013), JSmol and the Next-Generation Web-Based Representation of 3D Molecular Structure as Applied to Proteopedia. Isr. J. Chem., 53:207-216. doi:http://dx.doi.org/10.1002/ijch.201300024
- ↑ Herraez A. Biomolecules in the computer: Jmol to the rescue. Biochem Mol Biol Educ. 2006 Jul;34(4):255-61. doi: 10.1002/bmb.2006.494034042644. PMID:21638687 doi:10.1002/bmb.2006.494034042644