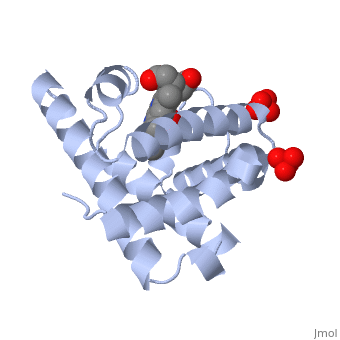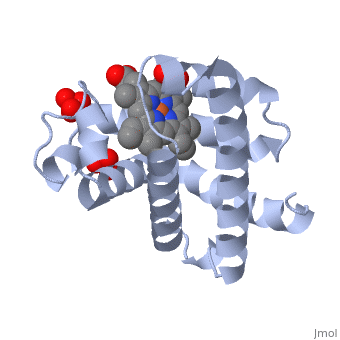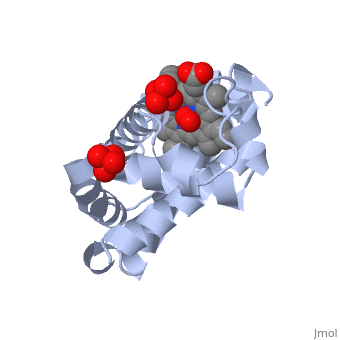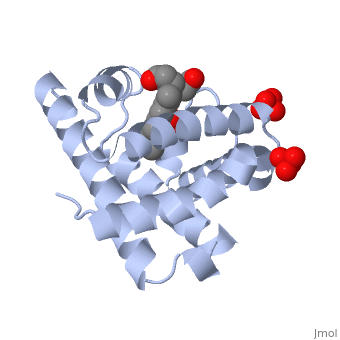Myoglobin
|
Caution: The text in this article and has not been updated to reflect what is actually available on this page. There is no zoom slider and no animate button. These were formerly present when an earlier version of Proteopedia supported Kinemages. A volunteer is needed to clean up and improve this article on a pedagogically and historically important protein. Green links are needed! Text referring to the now absent Kinemage is in italics. Eric Martz 01:09, 13 September 2014 (IDT) |
Myoglobin is a globular protein whose function is to store molecular oxygen. The overall of myoglobin is approximately disc-shaped with a diameter that is about twice its thickness. The overall fold of the protein is conserved, especially the core of the protein (shown in purple), but the sequence is more on the surface. The globin consists mostly of alpha helices shown in ; it has no beta sheets and its nonhelical segments mostly serve as links that connect the helices. Look down the barrel of some of the longer helices. Are they all straight? The eight structurally conserved alpha helices are labelled . The protein is colored as a N-->C rainbow in this view; the N terminus is blue, while the C terminus is red. The , and specifically the iron atom in the middle of the heme, is what binds oxygen in myoglobin. In this representation, the heme alone is shown in ball and stick form with its c, N and O atoms displayed as grey, blue, and red balls. The iron atom is shown in orange, and is in spacefill to better illustrate its interactions with the heme. The iron is bound by four nitrogen atoms found in the heme ring, as well as an from the protein chain. Which amino acid from the myoglobin protein binds to the iron? Notice that in the oxygenated state, the iron is in the plane of the heme ring. Note how the heme is almost completely enclosed by the globin. Which few chemical groups of the Heme are exposed to the solvent? (holding the mouse over the atoms reveals their identity.) Can you rationalize this exposure?
Turn on the "Main Chain" button to display the polypeptide backbone in white with its N and O atoms represented by blue and red balls. How closely does the ribbon follow the main chain? Click "Animate" until the Mb ribbon is gray. Turn off the "Mb Ribbon" button. What are the orientations of the main chain carbonyl groups and the amide N atoms relative to each other which allows formation of the H-bonds of the alpha helices (not drawn)? See also Molecular Playground/Myoglobin Myoglobin page in Spanish Myoglobin-Physeter-catodon-structure.
|
| ||||||||||
Former exercise in large part by John H. Connor (present address: Department of Microbiology, Boston University School of Medicine, 850 Harrison Ave, Boston, MA, 02118, USA). Revised by Ann Taylor
3D Structures of Myoglobin3D Structures of Myoglobin
Updated on 30-March-2015
Myoglobin (Mb) is an oxygen binding protein found in muscle tissue. It contains a heme group. Metmyoglobin (MMb) is the oxidized form of myoglobin.
((3qm5, 3qm6 – btMb – blackfin tuna
- Mb mutants uncomplexed
- 3m38, 3m39, 3m3a, 3m3b, 3k9z, 3h57, 3h58, 2e2y, 2ef2, 2oh8, 2oh9, 2oha, 2ohb, 2blh, 2bli, 1h1x, 1co8, 1n9h, 1n9i, 1n9x, 1naz, 1f63, 1f65, 1dti, 1co9, 1cp0, 1cp5, 1cpw, 1ch1, 1ch2, 1ch3, 1ch5, 1ch7, 1ch9, 1cik, 1cio, 1ofk, 1ofj, 102m, 1obm, 2mbw, 1tes, 1irc, 1mti, 1mtj, 1mtk, 1mlf, 1mlg, 1mlh, 1mlj, 1mlk, 1mll, 1mlm, 1mlo, 1mlr, 1mln, 1mls, 2mga, 2mgb, 2mgc, 2mgd, 2mge, 2mgf, 2mgg, 2mgh, 2mgi, 2mgj, 2mgk, 2mgl, 2mgm, 2spl, 2spm, 2spn, 2spo, 1fcs, 1j52, 1lue, 1o16, 3ogb, 3obd, 3ock, 3sdn, 3o89, 4fwx, 4fwz, 4it8 - SwMb (mutant)
- 2bwh - SwMb (mutant) – Laue
- 3hc9, 3hen, 3heo, 3hep, 1nz2, 1nz3, 1rse, 1abs, 1xch, 1hrm, 1yma, 3rj6 - hoMb (mutant) – horse
- 2vlx , 2vly, 2vlz, 2vm0 – hoMb fragment
- 1dm1 – AlMb (mutant)
- 2mm1, 3rgk – hMb (mutant) – human
- 3m38, 3m39, 3m3a, 3m3b, 3k9z, 3h57, 3h58, 2e2y, 2ef2, 2oh8, 2oh9, 2oha, 2ohb, 2blh, 2bli, 1h1x, 1co8, 1n9h, 1n9i, 1n9x, 1naz, 1f63, 1f65, 1dti, 1co9, 1cp0, 1cp5, 1cpw, 1ch1, 1ch2, 1ch3, 1ch5, 1ch7, 1ch9, 1cik, 1cio, 1ofk, 1ofj, 102m, 1obm, 2mbw, 1tes, 1irc, 1mti, 1mtj, 1mtk, 1mlf, 1mlg, 1mlh, 1mlj, 1mlk, 1mll, 1mlm, 1mlo, 1mlr, 1mln, 1mls, 2mga, 2mgb, 2mgc, 2mgd, 2mge, 2mgf, 2mgg, 2mgh, 2mgi, 2mgj, 2mgk, 2mgl, 2mgm, 2spl, 2spm, 2spn, 2spo, 1fcs, 1j52, 1lue, 1o16, 3ogb, 3obd, 3ock, 3sdn, 3o89, 4fwx, 4fwz, 4it8 - SwMb (mutant)
- Mb containing a non-Fe protoporphyrin
- 1yog, 1yoh, 1yoi, 1myz – SwMb+Co protoporphyrin
- 3mn0 – SwMb (mutant)+Cu protoporphyrin+cyanide
- 4mxk, 4mxl - SwMb (mutant)+Zn protoporphyrin
- 1j3f – SwMb+Cr salophen
- 1ufj, 1ufp – SwMb (mutant)+Fe salophen
- 2o58, 2o5b, 3wi8 – hoMb+Mn protoporphyrin
- 2o5l - hoMb+Mn protoporphyrin +methanol
- 2o5m - hoMb+Mn protoporphyrin +azide
- 2o5o, 2o5q - hoMb+Mn protoporphyrin +NO2
- 2o5s - hoMb+Co protoporphyrin +NO2
- 2o5t - hoMb+Co protoporphyrin
- 3rjn - hoMb+Zn deuteroporphyrin
- 1yog, 1yoh, 1yoi, 1myz – SwMb+Co protoporphyrin
- Mb+NO
- Mb+NO2
- Mb+O2
- Mb+CO
- 2bw9, 1mz0 - SwMb (mutant)+CO – Laue
- 2g0r, 2g0s, 2g0v, 2g0x, 2g0z, 2g10, 2g11, 2g12, 2g14, 2blj, 1dxc, 1dxd, 1do1, 1do3, 1do4, 1do7, 1abs, 1mcy, 1mbc, 3nml – SwMb (mutant)+CO
- 1bzr , 1a6g, 1ajg, 1ajh, 1spe, 1mym, 1mbc – SwMb+CO
- 1myf - SwMb+CO – NMR
- 2mb5 - SwMb+CO – Neutron
- 1dwr, 1dws, 1dwt – hoMb+CO
- 1mdn, 1m6c – pMb (mutant)+CO
- 1mwc, 1yca – pMb+CO
- 3qm7 – btMb + CO
- 2bw9, 1mz0 - SwMb (mutant)+CO – Laue
- Mb+OH
- Mb+cyano compounds
- 2jho – SwMb cyanoMet
- 3qm8 - btMb cyanoMet
- 1ebc, 2cmm - SwMb+cyanide
- 1iop – SwMb+cyanide+hemin
- 108m, 101m, 104m, 105m, 2myc, 2myd, 2mya, 2mye, 2myb – SwMb+isocyanides
- 103m, 107m, 111m, 106m, 109m, 110m, 112m – SwMb (mutant) +isocyanides
- 3ba2 – hoMb+cyanide
- 2fal – AlMb+cyanide
- 2fam – AlMb+thiocyanate
- 1lht – stMb+cyanide
- 1emy – Mb+cyanide – asian elephant
- 2jho – SwMb cyanoMet
- Mb + N3
- 3qm9 – btMb + N3
- MMb
- Other complexes of Mb
- 3a2g – SwMb (mutant)+fluorescein
- 2w6x, 2w6y - SwMb (mutant)+Xe
- 2w6w - SwMb +Xe
- 2eb8, 2eb9 – SwMb+Cu
- 4fwy - SwMb (mutant)+ Cu
- 3ase – SwMb + RuO3
- 2ekt, 2eku – SwMb+methyl-depropionatehemin
- 2d6c – SwMb+porphycene
- 1swm, 1mbi – SwMb+imidazole
- 3u3e – SwMb + phenol
- 4h07 - SwMb (mutant)+ phenol
- 4h0b - SwMb (mutant)+ DMSO
- 1wvp – SwMb+tetrazolyl histidine
- 2evk – SwMb (mutant)+acetic acid
- 2evp - SwMb (mutant)+mercaptoethanol
- 1v9q - SwMb (mutant)+Mn salophen derivative
- 1dtm - SwMb (mutant)+4-methylimidazole
- 2nsr – hoMb+nitro methane
- 1npg – hoMb+nitrosoethane
- 2nss - hoMb+nitro benzene
- 1azi - hoMb+azide
- 1bje – hoMb (mutant)+azide
- 1nz4 – hoMb (mutant)+Cd
- 1nz5 - hoMb (mutant)+Mn
- 2in4 - hoMb+Zn-DME
- 1gjn – hoMb+hydroxide
- 2nrm – BtMb S-nitrosylated
- 5mba – AlMb+azide
- 1mni – pMb+imidazole
- 3qma - btMb+imidazole
- 3a2g – SwMb (mutant)+fluorescein
- apo-Mb