Rossmann fold
Rossmann fold is a super-secondary structure that is characterized by an alternating motif of beta-strand-alpha helix-beta strand secondary structures. Hence this fold is also called a βαβ fold. The β-strands participate in the formation of a β-sheet. The βαβ fold structure is commonly observed in enzymes that have dinucleotide coenzymes, such as FAD, NAD and NADP.
HistoryHistory
In 1973 Rao and Rossmann reported that a βαβ fold super-secondary structure commonly appears in a variety of nucleotide binding proteins, such as lactate dehydrogenase and flavodoxin [1]. In later studies this common structure was named after the author Michael G. Rossmann as Rossmann fold.
The βαβ fold structure was initially characterized in dinucleotide FAD and NADH binding proteins. Schulz et al. examining FAD binding domains of four enzymes noted that the βαβ fold structure was associated with a specific consensus sequence of Gly-x-Gly-x-x-Gly at the region of the tight loop between the first β-strand the α-helix [2]. Wierenga et al. systematically examined more structures and derived rules for a fingerprint sequence named "ADP-binding βαβ fold" [3].
In 1989, Israel Hanukoglu found that NADPH binding βαβ fold in NADP dependent enzymes is characterized by a specific consensus sequence (briefly: Gly-x-Gly-x-x-Ala) that differs from the NADH binding site by one residue, with an alanine instead of the last glycine, and hypothesized that this single residue difference may determine the coenzyme specificity of the enzymes [4]. Richard Perham and his colleagues confirmed this hypothesis by site-directed mutagenesis of glutathione reductase and showed that coenzyme specificity could be re-engineered from NAD to NADP [5]. The structural significance of the Ala for Gly substitution in NADP binding site was revealed by analysis of the crystal structure of NADP-dependent adrenonodoxin-reductase [6].
Dinucleotides that bind to Rossmann foldDinucleotides that bind to Rossmann fold
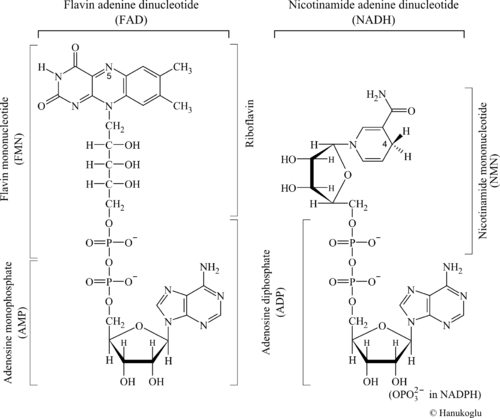
The βαβ fold is observed in numerous dinucleotide binding enzymes. The term "dinucleotide" may have two meanings: It may refer to an oligomer of two nucleotides such as A-G. In the present context, the term dinucleotide refers to a coenzyme that contains two distinct nucleotides. To emphasize the common structural aspects, the structures of two dinucleotides, FAD, NADH are shown in Figure 1.
Note that both dinucleotides share at their base the common structure of adenosine diphosphate (ADP). The structure of FAD can be viewed either as a hybrid of AMP+FMN or as ADP+Riboflavin (see figure).
NAD(P) is a two electron acceptor and donates the two electrons to FAD. The transfer of electrons takes place from C4 of NAD(P) to N5 of FAD. Both of these atoms are marked by their respective number in Figure 1.
Contact region between Rossmann fold and FADContact region between Rossmann fold and FAD
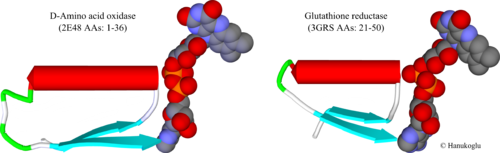
In dinucleotide binding flavoproteins, FAD binding Rossmann fold is commonly located close to the amino terminus of the protein. Figure 2 shows the first Rossmann fold of two flavoproteins, D-amino acid oxidase (2e48)[7] and glutathione reductase (3GRS)[8]. In both enzymes, the first β-strand is followed by a tight loop that is connected to the N-terminal of the helix. The two highly conserved Gly residues in the consensus sequence are located in this turn to allow the sharp bending of the chain. At the end of the helix there is a wider turn that is followed by the second beta strand that runs parallel to the first strand.
FAD structure is shown in CPK format. The atoms can be identified by their colors: C: grey; O: red, P: orange and N: purple. The turn at β-α border is in contact with the negatively charged oxygens (red colored) of the two phosphate (orange colored) groups.
To see the locations of the conserved glycines in the consensus sequence, click the following residues
Gly-x-Gly-x-x-Gly
Contact region between Rossmann fold and NAD(P)Contact region between Rossmann fold and NAD(P)
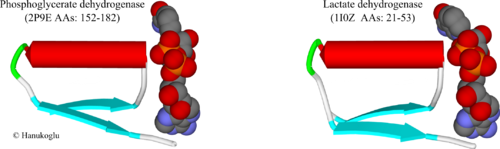
Figure 3 shows the Rossmann fold of two NAD binding proteins, 3-phosphoglycerate dehydrogenase (2P9E)[9] and lactate dehydrogenase (1I0Z)[10]. In enzymes that have just an NAD binding site, the site may be close to the N terminus of the protein as in lactate dehydrogenase. In flavoproteins that bind two dinucleotides, such as glutathione reductase and adrenodoxin reductase [4], the NAD(P) binding site appears in the middle of the protein.
The βαβ fold has a structure similar to the fold shown above for FAD. For both enzymes, the first β-strand is followed by a tight turn that is connected to the N-terminal of the helix. The same Gly-x-Gly-x-x-Gly consensus sequence appears at the turn between the first strand and the helix. Again, similar to FAD site, the turn region is in contact with the negatively charged oxygens (red colored) of the two phosphate (orange colored) groups.
Extension of the beta sheet by additional strandsExtension of the beta sheet by additional strands

In many (but not all) proteins with βαβ fold, the β-strands may be part of a larger β-sheet with up to seven β-strands. Figure 4 shows five strands forming a β-sheet in phosphoglycerate dehydrogenase (2P9E). Note that the segment connecting the second strand to the third is in coiled confirmation and not helical. Whereas the subsequent connections between strands include α-helix segments.
Evolutionary origin of the βαβ foldEvolutionary origin of the βαβ fold
A myriad of proteins include the βαβ Rossmann fold. Many of these proteins can be grouped in a hierarchy of families based on their sequence similarities [11],[12],[13]. Yet, many of these families do not show any significant sequence homology across families.
The observation that the βαβ structure and its consensus sequence is observed in many seemingly unrelated proteins raises the question whether the origin of the βαβ fold of all these proteins is a common ancestral sequence. Alternatively, there is also a possibility that this structure emerged in different proteins independently.
The basic nucleus of the βαβ fold is about 30 residues. In many proteins that do not share any significant sequence homology, there exists an extensive tertiary structural homology beyond this 30 residue segment, particularly in specific domains that bind dinucleotides. Therefore, the probability that this type of extensive structural homology evolved independently is very low. Thus, most likely βαβ fold represents an ancient structure that left its vestige in numerous proteins. Certainly, this conclusion does not exclude the possibility of independent convergent evolution.
ReferencesReferences
- ↑ Rao ST, Rossmann MG. Comparison of super-secondary structures in proteins. J Mol Biol. 1973 May 15;76(2):241-56. PMID:4737475
- ↑ Schulz GE, Schirmer RH, Pai EF. FAD-binding site of glutathione reductase. J Mol Biol. 1982 Sep 15;160(2):287-308. PMID:7175934 doi:http://dx.doi.org/10.1016/0022-2836(82)90177-2
- ↑ Wierenga RK, Terpstra P, Hol WG. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986 Jan 5;187(1):101-7. PMID:3959077 doi:http://dx.doi.org/10.1016/0022-2836(86)90409-2
- ↑ 4.0 4.1 Hanukoglu I, Gutfinger T. cDNA sequence of adrenodoxin reductase. Identification of NADP-binding sites in oxidoreductases. Eur J Biochem. 1989 Mar 15;180(2):479-84. PMID:2924777
- ↑ Scrutton NS, Berry A, Perham RN. Redesign of the coenzyme specificity of a dehydrogenase by protein engineering. Nature. 1990 Jan 4;343(6253):38-43. PMID:2296288 doi:http://dx.doi.org/10.1038/343038a0
- ↑ Ziegler GA, Vonrhein C, Hanukoglu I, Schulz GE. The structure of adrenodoxin reductase of mitochondrial P450 systems: electron transfer for steroid biosynthesis. J Mol Biol. 1999 Jun 18;289(4):981-90. PMID:10369776 doi:10.1006/jmbi.1999.2807
- ↑ Kawazoe T, Tsuge H, Imagawa T, Aki K, Kuramitsu S, Fukui K. Structural basis of D-DOPA oxidation by D-amino acid oxidase: alternative pathway for dopamine biosynthesis. Biochem Biophys Res Commun. 2007 Apr 6;355(2):385-91. Epub 2007 Feb 8. PMID:17303072 doi:10.1016/j.bbrc.2007.01.181
- ↑ Karplus PA, Schulz GE. Refined structure of glutathione reductase at 1.54 A resolution. J Mol Biol. 1987 Jun 5;195(3):701-29. PMID:3656429
- ↑ Dey S, Hu Z, Xu XL, Sacchettini JC, Grant GA. The effect of hinge mutations on effector binding and domain rotation in Escherichia coli D-3-phosphoglycerate dehydrogenase. J Biol Chem. 2007 Jun 22;282(25):18418-26. Epub 2007 Apr 24. PMID:17459882 doi:10.1074/jbc.M701174200
- ↑ Read JA, Winter VJ, Eszes CM, Sessions RB, Brady RL. Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase. Proteins. 2001 May 1;43(2):175-85. PMID:11276087
- ↑ Dym O, Eisenberg D. Sequence-structure analysis of FAD-containing proteins. Protein Sci. 2001 Sep;10(9):1712-28. PMID:11514662 doi:10.1110/ps.12801
- ↑ Lesk AM. NAD-binding domains of dehydrogenases. Curr Opin Struct Biol. 1995 Dec;5(6):775-83. PMID:8749365
- ↑ Ojha S, Meng EC, Babbitt PC. Evolution of function in the "two dinucleotide binding domains" flavoproteins. PLoS Comput Biol. 2007 Jul;3(7):e121. PMID:17658942 doi:10.1371/journal.pcbi.0030121