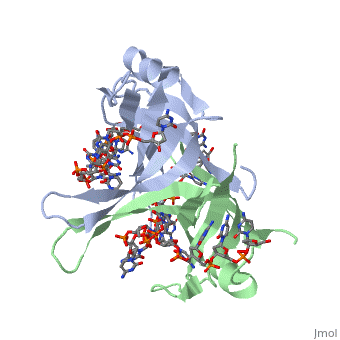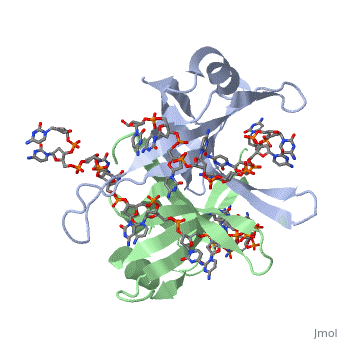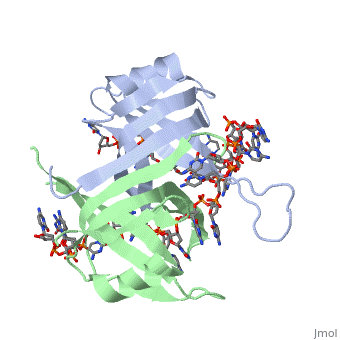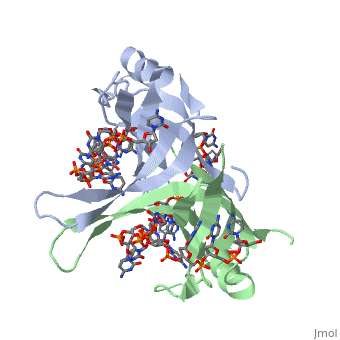
Single stranded binding protein
Single-stranded DNA-binding protein, or SSB, binds to single-stranded regions of DNA in order to prevent premature annealing, to protect the single-stranded DNA from being digested by nucleases, and to remove secondary structure from the DNA to allow other enzymes to function effectively upon it. Single-stranded DNA is produced during all aspects of DNA metabolism: replication, recombination and repair. As well as stabilizing this single-stranded DNA, SSB proteins bind to and modulate the function of numerous proteins involved in all of these processes.
SSB proteins have been identified in organisms from viruses to humans. The only organisms known to lack them are Thermoproteales, a group of extremophile archaea, where they have been displaced by the protein ThermoDBP. While many phage and viral SSBs function as monomers and eukaryotes encode heterotrimeric RPA (Replication Protein A), the best characterized SSB is that from the bacteria E. coli which, like most bacterial SSBs exists as a tetramer. Active E. coli SSB is composed of four identical 19 kDa subunits. Binding of single-stranded DNA to the tetramer can occur in different "modes", with SSB occupying different numbers of DNA bases depending on a number of factors, including salt concentration. For example, the (SSB)65 binding mode, in which approximately 65 nucleotides of DNA wrap around the SSB tetramer and contact all four of its subunits, is favoured at high salt concentrations in vitro. At lower salt concentrations, the (SSB)35 binding mode, in which about 35 nucleotides bind to only two of the SSB subunits, tends to form. Further work is required to elucidate the functions of the various binding modes in vivo.
Sandbox Single Stranded DNA-Binding Protein (SSB)Sandbox Single Stranded DNA-Binding Protein (SSB)
IntroductionThe single stranded DNA binding protein (SSB) of E. coli plays an important role in three aspects of DNA metabolism – namely in replication, repair and recombination. During DNA replication, SSB molecules bind to the newly separated DNA strands, keeping the strands separated by holding them in place so that each strand can serve as a template [1].
|
| ||||||||||
StructureSSB consists is a homotetramer that has a DNA binding domain which binds to a single strand of DNA. The tetramers consist of α-helices, β-sheets, and random coils. Each subunit contains an and several . The secondary structure also includes a NH2 terminus, which consists of multiple positively charged amino acids. The COOH terminus includes many acidic amino acids. Binding Interactions in the Active Siteis an important DNA binding site. It has been shown to be the site for cross-linking. Tryptophan and Lysine residues are important in binding as well. Treatments resulting in modification of arginine, cysteine, or tyrosine residues had no effect on binding of SSB to DNA, whereas modification of either lysine residues (with acetic anhydride) or tryptophan residues (with N-bromosuccinimide) led to complete loss of binding activity [2]. The two tryptophan residues involved in DNA binding are Trp40 and Trp54, which was determined by mutagenesis. One more binding site was determined by site-specific mutagenesis. When His55 is substituted with Leu it decreases binding affinity. All of these residues are found in a hydrophobic region, which is suitable for nucleotide base interactions. SSB-Protein InteractionsIt is believed that Gly15 may play an important role in binding the RecA protein. Mutations in Gly15 have extreme effects on recombinational repair. SSB has also been thought to bind with exonuclease I, DNA polymerase II, and a protein n, which is used to help synthesize RNA primers for the lagging strand. |
| ||||||||||
|
Binding Interactions between DNA and SSB of E. coliPhe60 is an important DNA binding site. It has been shown to be the site for cross-linking. and Lysine residues are important in binding as well. Treatments resulting in modification of arginine, cysteine, or tyrosine residues had no effect on binding of SSB to DNA, whereas modification of either lysine residues (with acetic anhydride) or tryptophan residues (with N-bromosuccinimide) led to complete loss of binding activity [3]. The two tryptophan residues involved in DNA binding are Trp40 and Trp54, which was determined by mutagenesis. One more binding site was determined by site-specific mutagenesis. When His55 is substituted with Leu it decreases binding affinity. All of these residues are found in a hydrophobic region, which is suitable for nucleotide base interactions. SSB-Protein InteractionsIt is believed that may play an important role in binding the RecA protein. Mutations in Gly15 have extreme effects on recombinational repair. SSB has also been thought to bind with exonuclease I, DNA polymerase II, and a protein n, which is used to help synthesize RNA primers for the lagging strand.
| ||||||||||
See AlsoSee Also
ReferencesReferences
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220
- ↑ Meyer RR, Laine PS. The single-stranded DNA-binding protein of Escherichia coli. Microbiol Rev. 1990 Dec;54(4):342-80. PMID:2087220