Antibody
Antibodies, also known as Immunoglobulins (Ig) are gamma globulin proteins, primarily found in the blood of vertebrates. These glycoproteins serve as a critical component of the immune system when the host fails to activate alternative compliment pathways or phagocytic cells in response to invading microorganisms or other antigens. The incredible specificity with which immunoglobulins bind to an antigen is based upon structural complementarity between the antigen and antibody and . It is this specificity that has made a critical component in laboratory and medical research. See more in Monoclonal Antibody. For Anti-HIV Fab see Human Fab PG16.
| |||||||||
| Crystal Structure of the Intact Human IGG B12: A Template for a Potential HIV Vaccine, 1hzh | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Ligands: | , , , , | ||||||||
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
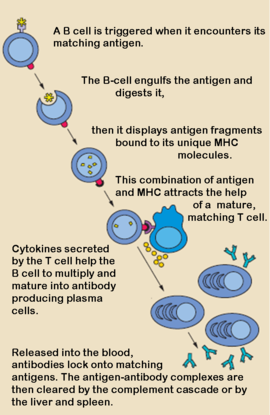
Cellular Basis of Antibody ProductionCellular Basis of Antibody Production
When a foreign antigen binds to a B-lymphocyte (B-cell), it activates the B-cell, and upon stimulation by helper T-cells, undergoes clonal proliferation and B-cell maturation into antibody forming plasma cells. Each plasma cell is programmed to make an antibody of a single specificity, which it releases into the blood. [1] Once in the blood, antibodies aid the humoral immune system in three predominant ways: They coat foreign pathogens preventing them from entering healthy cells or disrupting antigen function; they coat pathogens, stimulating their removal via opsonization by phagocytes; and they trigger destruction of pathogens by stimulating the complement pathway or by Antibody Dependent Cell-mediated Cytotoxicity, among other immune responses. [2] [3] All of these functions rely heavily on accurate antigen binding and communication with other immune effector cells. The amazing specificity antibodies operate with is made possible by the physical structure of the antibody, which appears simplistic, but contains several levels of additional complexity.
Structure of the ImmunoglobulinStructure of the Immunoglobulin
Template:STRUCTURE 1igt The basic functional unit of an antibody is an immunoglobulin monomer, but antibodies secreted from plasma cells are typically dimeric with occasional higher order structures. Typical secreted antibodies have a basic four-peptide structure of two identical and two identical joined together by interchain , forming a “Y” shaped molecule. The disulfide bonds are positioned within a flexible region called the , which seperates the lobes of the antibody from one another and provides ample flexibility to bind antigens effectively. [1] Each domain (2 heavy and 2 light) contain between 70-110 amino acids and are classified into different categories according to size and function. [4] Both domains, heavy and light, contain variable and constant regions that are crucial to antibody function. [5]
Heavy ChainHeavy Chain
There are five types of immunoglobulin heavy chains, in mammals, α, δ, ε, γ, and μ, and give rise to the five unique classes or isotypes of antibodies, IgA, IgD, IgE, IgG, and IgM, which differ in size and composition. Each has a and . The constant region is identical in all antibodies of the same isotype, but differ in antibodies of different isotypes; i.e. all IgA have the same sequence in their heavy chain constant region, but these constant regions differ between IgA and IgD, etc. [6] The α, δ, and γ heavy chains have a constant region composed of while heavy chains ε and μ contain four. The variable region of the heavy chain in antibodies is different for all antibodies created by different B-cells. [7]
Light ChainLight Chain
Every antibody contains two that are identical to each other. There are two types of immunoglobulin light chains in mammals, labeled lambda and kappa, with only one represented in each antibody. Each light chain has one followed by one , with a total length of about 215 amino acids. [1]
The Regions: Fab, Fv, CDR, and Fc.The Regions: Fab, Fv, CDR, and Fc.
The immunoglobulin can be broken down into regions, each serving a different purpose:
Variable RegionsVariable Regions
The (Fragment, Antigen Binding region) is composed of one constant and one variable domain from each heavy and light chain of the antibody. It is the part of the antibody that gives it its famous “Y” shape.[8] Held within the Fab region is the variable domain, also known as the Fv region.[9] Within the Fv region lie positioned at one end of the variable domain where they form parts of the Beta-turn loops and are clustered close to each other in space. The clustering of the hypervariable loops at the tips of the variable regions where the antigen-binding site is located makes them perfect candidates for antigen recognition. [1]The sequence heterogeneity of the three heavy and three light chain hypervariable loops creates significant antigen specificity diversity through variations in the binding surface nature and shape. Each hypervariable region can be viewed as an independent structure contributing to the complementarity of the biding site and antigen and is often referred to as a complementarity determining region (CDR). [10]
Constant RegionsConstant Regions
The remaining part of the antibody, namely the , does not play a role in binding the antigen, but rather is responsible for modulating the immune systems response to the formation of an antibody-antigen complex. The Fragment Crystallizable (Fc) region is composed of two heavy chain constant regions that are isotype specific. [11] Antibodies are glycoproteins because of at conserved positions in their Fc regions. This glycosylation is a critical component determing the rate of antibody clearance form the body.[12] Once an antibody binds to an antigen, the Fc region binds to Fc receptors, among other proteins, to mediate a host of different physiological responses ranging from oposonization, to degranulation of mast cells, to the release of cytokines and cytotoxic molecules, etc. resulting in the destruction of the pathogen. [13] Depending on the class of antibody, as dictated by the identity of the Fc region, the antibody half-life and distribution throughout the body varies. Further, since Fc receptors are antibody isotype specific, the type of immune response is dependent on the type of Fc region on the immunoglobulin, allowing for different immune responses to the same pathogen if necessary.[14] See table for brief characterization of Immunoglobulin isotypes:
| Immunoglobulin Classes and Function | |
|---|---|
| Class | Function and Oligomeric State[1] |
| IgG | Dimeric - The most abundant Ig in the extravascular fluids. Neutralizes toxins and combats microorganisms by activating the compliment system and facilitating the binding of phagocytic cells. |
| IgA | Dimeric - Is the major Ig in seromucous secretions, where it serves to defend the external body surfaces. |
| IgM | Pentameric – It is an intravascular antibody and is produced very early in the immune response. Due to it high oligomeric state, it is extremely effective as a bacterial agglutinator and mediator of complement-dependent cytolysis, making it a powerful first-line defense against bacterial pathogens. |
| IgD | Dimeric - It is present on the lymphocyte and functions together with IgM as the antigen receptor on naïve B-cells. |
| IgE | It binds to mast cells and upon contact with antigen, leads to local recruitment of antimicrobial agents via degranulation of the mast cell and release of inflammatory mediators. IgE is important for certain kinds of parasitic infections and is responsible for the symptoms of atopic allergies like eczema and asthma. |
A model of the IgG molecule is present in the figure which indicates the spatial disposition and interaction of the domains in IgG. As Dr. Ivan Roitt writes in Essential Immunolgy, “To enable the Fab arms to have the freedom to move and twist so that they can align their hypervariable regions with the antigenic sites on large immobile carriers, and to permit the Fc structures to adjust spatially in order to trigger their effector functions, it is desirable for IgG to have a high degree of flexibility. And it has just that. Structural analysis shows that the Fab can ‘elbow-bend’ at its V-C junction and twist about the hinge, which itself can more properly be described as a loose thether, allowing the Fab and the Fc to drift relative to each other with remarkable suppleness. It could be said that movements like that make it a very sexy molecule!” [1]
|
Antibody DiversityAntibody Diversity
Considering the nearly infinite number of possible antigens that can invade the body, the immune system had to develop a method for accurately targeting each one of these compounds, ranging from small molecules, to stray proteins, to viruses capable of infecting cells. The antibody was the immune systems response to this problem. It has been estimated that humans generate about 10^10 different antigens, each capable of binding a unique epitope of an antigen. Since antibodies are proteins, and proteins are controlled by the genes from which they are transcribed, a clever system of gene shuffling and manipulations developed to enable the immune system to create a huge repertoire of antibodies from a limited number of genes. [15] The variable region of each immunoglobulin chain is encoded in several pieces known as gene segments. For heavy chains, these segments are called the variable (V), diversity (D), and joining (J) segments. (Only V and J exist for light chains) 50 V segments, 25 D segments, and 6 J segments exist and are randomly arranged and rearranged in the genome in a process called V(D)J recombination. Each B-cell is programmed to produce antibodies of a single V(D)J recombination order.
Additional diversity is created by the proteins RAG-1 and RAG-2 which introduce the double stranded breaks between V, D, and J segments to allow recombination. At this stage, nucleotides can either be deleted or inserted between adjoining segments before being ligated together. [1] This dramatically increases antibody diversity. Further diversity is created during B-cell proliferation when the variable chains undergo a high rate of point mutations in a process called somatic hypermutation, creating daughter cells of the original B-cell that are slightly different. The antibodies which bind the antigen with the highest affinity are selected for in a process called affinity maturation. [16][17] Isotype switching is also possible after activation of the B-cell by a mechanism called “class switch recombination” allowing different immunological responses to the same antigen bound by the same variable regions.[18] Through this clever system, tens of billions of different glycoprotein antibodies can be created from less than 100 genes, allowing antibodies to bind with exquisite precision. The discovery of antobdy diversity generation won Susumu Tonegawa the Nobel Prize in Medicine in 1987.
Antibody ApplicationsAntibody Applications
Detection of particular antibodies is very common in medical diagnostic testing. Numerous biochemical assays exist to detect whether antibodies for specific antigens are present in the blood or other bodily fluids such as antibodies against Lyme disease or HIV, etc. Another common medical test involving antibodies is blood type detection in which an individual’s blood is screened against anti-A and anti-B antibodies to determine the identity of that individual’s blood antigen type. [19]
Antibodies are also extremely powerful tools in the laboratory setting where they are commonly used in Western Blot to detect specific proteins in a sample [20]; flow cytometry, to differentiate cell types by their protein expression profiles; immunoprecipitation, to separate proteins from other compounds in a lysate and for cellular labeling. Numerous other examples exist. [21]
The last two decades have seen a dramatic increase in antibody based technologies both for the lab and medicine thanks to the invention of the monoclonal antiboy, a discovery that won Niels K. Jerne, Georges J.F. Köhler, César Milstein the Nobel Prize in Medicine in 1984. See: Monoclonal Antibody for additional information.
3D Structures of the Immunoglobulin3D Structures of the Immunoglobulin
Update June 2012
Humanized mouse antibody (hmFab) is a modified mFab which resembles more hFab.
FabFab
7fab - hFab - human
3o2v – hFab 1e9 (mutant)
3na9 – hFab 15
3naa, 3nab, 3nac, 3ncj - hFab 15 (mutant)
3qct – hFab anti-lysophosphatidic acid
3nfs – hFab commercial
3lrs, 3mme – hFab PG16
3hi5 – hFab AL-57
1vge – hFab TR1.9
1hkl – hFab catalytic
3f12 – hFab M2J1
8fab – hFab HIL
1opg - hFab OPG2
1om3 – hFab 2G12
1aqk – hFab B7-15A2
2hff – hFab CB2
2agj – hFab YVO
3ls5 – hFab anti-tetrahydrocannabinol
3hc0 – hFab BHA10
3hc3, 3hc4 - hFab BHA10 (mutant)
3g6a – hFab CNTO607
3dgg, 3dif – hFab OX108
3eyo, 3eyq – hFab 8F9
2aj3 – hFab M18
3fzu – hFab igG1
3aaz, 3gje – hFab
1jpt - hmFab D3H44
3mxv – mFab/hFab
2o5x - mFab/hFab 1E9-DB3
1ucb - mFab/hFab BR96
1f4w – mFab S-20-4
1aif – mFab 409.5.3
1ghf – mFab GH1002
2z91 – mFab 10C9
1ind – mFab CHA255
3bkc, 3bkm – mFab WO2
3iy0 – mFab 14
3pp3, 3pp4 – mFab GA101
3okm - mFab S25-39 igG1
6fab – mFab 36-71
2hkh – mFab M75
1ay1 – mFab TP7
1nbv – mFab BV04-01
1qbm – mFab E8B
1bbd - mFab 8F5
1igf – mFab B13I2
1for - mFab 17-IA
2gfb – mFab CNJ206
2rcs – mFab 48G7
2aju – mFab 7A1
1k6q – mFab D3
2fbj – mFab anti-galactan
1mcp – mFab anti-phosphocholine
1dqd – mFab HGR-2 F6
2ipt – mFab PFA1
1ct8 – mFab 7C8
1yeh, 1yec, 1yed, 1yee, 1eap - mFab catalytic
2iq9, 2iqa – mFab PFA2
2w60 – mFab ACC4
2w9d – mFab ICSM
3eot – mFab LAC031 (mutant)
3iu4 – mFab CHP3
1hq4 – mFab HA5-19A4
1q9k, 1q9l – mFab S25-2
1q9o - mFab S45-18
2z4q – mFab 528
1cr9 – mFab 3F4
1gig – mFab HC19
1uyw – mFab 4G2
1cgs, 2cgr – mFab NC6.8
1q0x – mFab 9B1
1ibg – mFab 40-50
3eo0 – mFab GC-1008
2op4 – mFab RS2-1G9
2q76 – mFab F10.6.6
2ojz – mFab ED10
2g60 – mFab M2
2dbl, 1dbj, 1dbk, 1dbm, 1dba – mFab DB3
1mrc – mFab JEL103
2gcy – mFab C25
1flr – mFab 4-4-20
1uz6 – mFab 291-2G3-A
1rfd – mFab M82G2
1t2q - mFab NNA7
15c8 – mFab 5C8
1ggb, 1ggc – mFab 50.1
1bm3 – mFab OPG2
2d03 – mFab NNA7 (mutant)
2eh7 – mFab KR127
2fat – mFab ATN-615
1c12 – mFab anti-traseolide
3cfj, 3cfk – hFab/mFab 34E4
1bbj - hFab/mFab B72.3
3mj8 – AhFab HL4E10 – Armenian hamster
3iy1 – rFab B – rat
1zan – rFab AD11
1t04 – hmFab HUZAF
2fgw – hmFab H52
1fvd, 1fve – hmFab 4D5
Anti-HIV FabAnti-HIV Fab
3lmj – hFab 21C anti-HIV
1rhh - hFab X5 anti-HIV
1rz7, 1rz8, 1rzf, 1rzg, 1rzi - hFab gp120-reactive anti-HIV
3mug - hFab PG16
3nz8 – mFab 7C8 anti-HIV
3o6k – mFab 11H6H1 anti-HIV
3ntc – mFab KD-247 anti-HIV
Fab: small molecule complexFab: small molecule complex
3o2w - hFab 1e9 (mutant) + transition state analog
3ra7 – mFab + digoxigenin – mouse
3okd, 3oke, 3okk, 3okl, 3okn, 3oko, 3hzk, 3hzm, 3hzv, 3hzy, 3pho, 3phq, 3hns, 3hnt, 3hnv - mFab + liposaccharide
3oau – hFab 2g12 (mutant) + mannose
1ikf – hFab R45 + cyclosporin
3oay, 3oaz, 3ob0, 1zls, 1zlu, 1zlv, 1zlw - hFab 2g12 + glycan
2jb5, 2jb6, 1y0l - hFab/mFab + hapten
2o5y, 2o5z - mFab/hFab 1E9-DB3 + steroid
3fo0 – hFab + hapten
1wcb, 2bmk, 1a0q, 1aj7, 1ine - mFab + hapten
3fo1, 3fo2 - hFab/mFab 13G5 (mutant) + hapten
3ls4 – mFab + tetrahydrocannabinol
1a4k, 1kno - mFab + transition state analog
1mfc, 1mfd, 1mfe – mFab + polysaccharide
1op3, 1op5 - hFab + polysaccharide
3eyv - hFab/mFab 13G5 (mutant) + hapten
2ntf - mFab RS2-1G9 + lactone analog
2ajs – mFab 7A1 + heptaethylene glycol
2ajv, 2ajx, 2ajy, 2ajz, 2ak1, 1riu, 1riv, 1qyg, 1q72 - mFab + cocaine derivative
1ynk, 1ynl, 1etz – mFab + sweetener
1yef – mFab D2.3 + substrate analog
1yeg - mFab D2.3 + product
1p7k – mFab + HEPES
1q0y – mFab 9B1 + morphine
1q9q, 1q9r, 1q9t, 1q9v, 1q9w, 1f4x, 1f4y, 1mfa, 1mfb - mFab + carbohydrate
1mex – mFab 29G12 + benzoic acid derivative
1mrd, 1mre, 1mrf – mFab JEL103 + nucleotide
1fig – mFab 1F7 + bicarboxylic acid
1dbb – mFab DB3 + progesterone
1igj - mFab 26-10 + digoxin
4fab – mFab 4-4-20 + fluorescin
2mcp - mFab MCPC603 + phosphocholine
Fab: peptide complexFab: peptide complex
3e8u - mFab + BNP peptide
3hr5 – mFab + M1prime peptide
3eys, 3eyu - mFab + amyloid-β-related peptide
3ggw – mFab + carbohydrate-mimetic peptide
3cxd, 3dsf – mFab + osteopontin peptide
2zpk – mFab + proteinase-activated receptor peptide
3ifl, 3ifn, 3ifo, 3ifp - mFab + amyloid peptide
3h0t - hFab + hepcidin peptide
3eyf – hFab + cytomegalovirus peptide
2hfg, 2h9g – hFab + TNF receptor peptide
3csy - hFab + Ebola envelope glycoprotein peptide
2eh8 - mFab + PRES1 peptide
2brr – mFab + outer membrane protein peptide
1xgy – mFab + rhodopsin peptide
1pz5 – mFab SYA/J6 + peptide
1a3r – mFab + rhinovirus capsid peptide
1cu4 - mFab + prion protein peptide
1fpt – mFab + poliovirus peptide
2hh0 – hFab/mFab + prion protein peptide
1frg, 1him, 1hin, 1ifh - mFab + hemagglutinin peptide
1tet – mFab + cholera toxin peptide
3bky – hFab/mFab + CD20 peptide
Anti-HIV Fab: peptide complexAnti-HIV Fab: peptide complex
3egs, 3drt, 3drq, 3dro, 2fx7, 2fx8, 2fx9, 2cmr, 1tzg, 1tjg – hFab anti-HIV + gp41 peptide
3ghb, 3ghe, 3c2a - hFab anti-HIV + envelope glycoprotein peptide
3go1 - hFab anti-HIV + envelope glycoprotein gp160 peptide
3fn0 - hFab Z13E1 anti-HIV + peptide
2oqj – hFab 2G12 anti-HIV + peptide
2b0s, 2b1a, 2b1h - hFab anti-HIV + glycoprotein gp120 peptide
1nak, 2f58, 3f58, 1f58, 1acy - mFab anti-HIV + glycoprotein gp120 peptide
1ai1, 1ggi – mFab anti-HIV + V3 peptide
3o6l, 3o6m - mFab 11H6H1 anti-HIV + Tat peptide
Fab: protein complexFab: protein complex
3pjs, 3efd, 3eff – mFab + KcsA K+ channel
3r1g – hFab + beta-secretase 1
3raj – mFab HB7 + CD38
3o0r – mFab + nitric oxide reductase
3mac, 3ma9 – hFab 8062 anti-HIV + transmembrane glycoprotein
3pnw – hFab + Tudor domain-containing protein 3
3h42 - hFab LDLR + proprotein convertase subtilisin/kexin
3nh7 – hFab ABD1556 + bone morphogenetic protein receptor
3hi6 – hFab AL-57 + integrin
2vxs - hFab + interleukin
3b2u, 3b2v – hFab IMC-11F8 + EGFR
2r0k, 2r0l – hFab + HGFA
3ld8 – CmFab + bifunctional arginine demethylase – Cricetulus migratorius
3be1, 3n85, 1n8z - hFab + ERBB-2
3kr3 – hFab + IGF-II
3g6d – hFab CNTO607 + IL-13
3idx, 3idy - hFab B13 anti-HIV + gp120 core
2x7l – Fab anti-HIV + HIV REV
3gbm, 3gbn, 3lzf - hFab + hemagglutinin
3g6j – hFab + complement C3
2vxq – hFab + pollen allergen PHL
2wub, 3k2u – hFab 40 + hepatocyte growth factor activator
3grw - hFab + fibroblast growth factor receptor
3bdy, 2qr0, 2fjg, 2fjh – hFab + VEGF
1tzh, 1tzi - mFab + VEGF
3bqu – hFab 2F5 anti-HIV + mFab 3H6
2j6e, 1adq – hFab IGM + igG1 Fc
3dvg, 3dvn – hFab igG1 + ubiquitin
3bn9 – hFab E2 + membrane-type serine protease
2jix – hFab ABT-007 + erythropoietin receptor
1za3 - hFab YSD1 + TNF receptor
3l95 – Fab + NRR1
1uj3 – hFab HATR-5 + tissue factor
1jps - hmFab D3H44 + tissue factor
1ahw - mFab 5G9+ tissue factor
2w9e, 1tpx, 1tqb, 1tqc – mFAB + major prion protein
2oz4 – mFab + intercellular adhesion molecule
3eo1 - mFab GC-1008 + transforming growth factor β-3
3d9a, 1bql, 1fbi, 2iff, 1fdl, 3hfm – mFab HYHEL10 + lysozyme
1ic4, 1ic5, 1ic7 - mFab HYHEL10 (mutant) + lysozyme
2dtg – mFab 83 + insulin receptor
1yjd – mFab 5.11A1 + CD28
1sy6 – mFab OKT3 + CD3
1afv – mFab anti-HIV + capsid protein C
2b2x – rFab AQC2 + integrin
2r56 – Fab igE + β-lactoglobulin - bovine
3iu3 – Fab basiliximab + inerleukin-2 receptor
2r9h – mFab + exchange transporter ClCA
1nca, 1ncb, 1ncc, 1ncd – mFab + neuraminidase
1rjl – mFab + outer surface protein
1mhh – mFab + protein L (mutant)
1pg7 – mFab 6A6 + hmFab D3H44
1pkq – hFab/mFab 8-18C5 + myelin glycoprotein
2jel – mFab JEL42 + histidine-containing protein
1nsn – mFab N10 + nuclease
1rvf – mFab 17-IA + intact rhinovirus
1nfd – mFab H57 + T-cell receptor
1igc – mFab MOPC21 + streptococcal protein G
1iai – mFab idiotipic + mFab anti-idiotypic
3ivk, 2r8s – mFab + RNA
2fr4, 1xf2, 1xf3, 1xf4, 1i8m, 1cbv – mFab + DNA
1mhp – mFab + integrin
Fab: protein ternary complexFab: protein ternary complex
3ixx, 3ixy - mFab E53 + envelope glycoprotein + West Nile virus peptide
2wuc - hFab 40 + hepatocyte growth factor activator + inhibitor
3lqa - hFab 21C anti-HIV + CD4 + gp160
3jwd, 3jwo - hFab 48D anti-HIV + CD4 + gp120
3d85 - hFab 7G10 + IL-12 + IL-23
3gb7, 2p7t, 2h8p, 2hfe, 2atk - mFab + KcsA K+ channel + ion
3or6, 3or7, 3ogc, 3fb7, 3f7v, 3f7y, 3fb5, 3fb6, 3fb8, 3iga, 2itc, 2itd, 2nlj, 2bob, 2boc, 1s5h, 1r3i, 1r3j, 1r3k, 1r3l, 1k4c, 1k4d - mFab + KcsA K+ channel (mutant) + ion
2fd6 – mFab ATN-615 + urokinase-type plasminogen activator + urokinase plasminogen receptor
Full ImmunoglobulinFull Immunoglobulin
1hzh - hFab IgG B12
Fc FragmentsFc Fragments
1f6a - hFc IgE + high-affinity receptor Fc (ε) RI (α)
1e4k - hFc Igg1 + Fc-γ-Riii Complex
1fp5 - hFc IgE C ε 3-C ε 4
ReferencesReferences
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 Roit, I. M. Roit's Essential Immunology. Oxford: Blackwell Science Ltd., 1997.
- ↑ Parker DC. T cell-dependent B cell activation. Annu Rev Immunol. 1993;11:331-60. PMID:8476565 doi:http://dx.doi.org/10.1146/annurev.iy.11.040193.001555
- ↑ Rus H, Cudrici C, Niculescu F. The role of the complement system in innate immunity. Immunol Res. 2005;33(2):103-12. PMID:16234578 doi:10.1385/IR:33:2:103
- ↑ Roux KH. Immunoglobulin structure and function as revealed by electron microscopy. Int Arch Allergy Immunol. 1999 Oct;120(2):85-99. PMID:10545762
- ↑ Putnam FW, Liu YS, Low TL. Primary structure of a human IgA1 immunoglobulin. IV. Streptococcal IgA1 protease, digestion, Fab and Fc fragments, and the complete amino acid sequence of the alpha 1 heavy chain. J Biol Chem. 1979 Apr 25;254(8):2865-74. PMID:107164
- ↑ Woof JM, Burton DR. Human antibody-Fc receptor interactions illuminated by crystal structures. Nat Rev Immunol. 2004 Feb;4(2):89-99. PMID:15040582 doi:10.1038/nri1266
- ↑ Putnam FW, Liu YS, Low TL. Primary structure of a human IgA1 immunoglobulin. IV. Streptococcal IgA1 protease, digestion, Fab and Fc fragments, and the complete amino acid sequence of the alpha 1 heavy chain. J Biol Chem. 1979 Apr 25;254(8):2865-74. PMID:107164
- ↑ Harris LJ, Larson SB, Hasel KW, McPherson A. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry. 1997 Feb 18;36(7):1581-97. PMID:9048542 doi:http://dx.doi.org/10.1021/bi962514+
- ↑ Hochman J, Inbar D, Givol D. An active antibody fragment (Fv) composed of the variable portions of heavy and light chains. Biochemistry. 1973 Mar 13;12(6):1130-5. PMID:4569769
- ↑ Putnam FW, Liu YS, Low TL. Primary structure of a human IgA1 immunoglobulin. IV. Streptococcal IgA1 protease, digestion, Fab and Fc fragments, and the complete amino acid sequence of the alpha 1 heavy chain. J Biol Chem. 1979 Apr 25;254(8):2865-74. PMID:107164
- ↑ Woof JM, Burton DR. Human antibody-Fc receptor interactions illuminated by crystal structures. Nat Rev Immunol. 2004 Feb;4(2):89-99. PMID:15040582 doi:10.1038/nri1266
- ↑ Wright A, Morrison SL. Effect of glycosylation on antibody function: implications for genetic engineering. Trends Biotechnol. 1997 Jan;15(1):26-32. PMID:9032990 doi:10.1016/S0167-7799(96)10062-7
- ↑ Heyman B. Complement and Fc-receptors in regulation of the antibody response. Immunol Lett. 1996 Dec;54(2-3):195-9. PMID:9052877
- ↑ Ravetch JV, Bolland S. IgG Fc receptors. Annu Rev Immunol. 2001;19:275-90. PMID:11244038 doi:19/1/275
- ↑ Fanning LJ, Connor AM, Wu GE. Development of the immunoglobulin repertoire. Clin Immunol Immunopathol. 1996 Apr;79(1):1-14. PMID:8612345
- ↑ Diaz M, Casali P. Somatic immunoglobulin hypermutation. Curr Opin Immunol. 2002 Apr;14(2):235-40. PMID:11869898
- ↑ Borghesi L, Milcarek C. From B cell to plasma cell: regulation of V(D)J recombination and antibody secretion. Immunol Res. 2006;36(1-3):27-32. PMID:17337763 doi:10.1385/IR:36:1:27
- ↑ Durandy A. Activation-induced cytidine deaminase: a dual role in class-switch recombination and somatic hypermutation. Eur J Immunol. 2003 Aug;33(8):2069-73. PMID:12884279 doi:10.1002/eji.200324133
- ↑ CHOWN B, LEWIS M, KAITA K. A new Kell blood-group phenotype. Nature. 1957 Oct 5;180(4588):711. PMID:13477267
- ↑ Burnette WN. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195-203. PMID:6266278
- ↑ Brehm-Stecher BF, Johnson EA. Single-cell microbiology: tools, technologies, and applications. Microbiol Mol Biol Rev. 2004 Sep;68(3):538-59. PMID:15353569 doi:10.1128/MMBR.68.3.538-559.2004
Additional PagesAdditional Pages
See AlsoSee Also
- Variable Lymphocyte Receptors
- Antibody at High school teachers' resources, where you will find tutorials on antibody structure.
- Antibodies at Wikipedia.
