Pho4 bHLH Protein

| |||||||||
| 1a0a, resolution 2.80Å () | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
| |||||||||
| Resources: | FirstGlance, OCA, RCSB, PDBsum | ||||||||
| Coordinates: | save as pdb, mmCIF, xml | ||||||||
PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEXPHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX
IntroductionIntroduction
The complete nucleotide sequence of Saccharomyces cerevisiae chromosome VI (270 kb) has revealed that it contains 129 predicted or known genes (300 bp or longer).(1)
One of them is PHO4 gene, which positively controls the expression of phosphatase genes, has been isolated by complementation of a pho4 mutation. The isolated DNA directed integration at the chromosomal PHO4 locus. The nucleotide sequence of PHO4 has a coding region of 930 nucleotides, flanked by sequences with typical transcription initiation and termination signals. The 5' region has characteristics of low-expression promoters and carries several uncommon elements, whose significance is not known. The predicted primary structure of the PHO4 protein, of 309 residues, does not show sequence elements typical of DNA-binding proteins. The transcription of PHO4 is independent of inorganic phosphate. Like other regulatory genes, PHO4 is transcribed at a very low level and the translation of its message uses preferentially several codons which are not employed for highly expressed genes. (2)
The PHO4 gene encodes a positive regulatory factor involved in regulating transcription of various genes in the phosphatase regulon of Saccharomyces cerevisiae. Besides its own coding region, the 1.8-kilobase PHO4 transcript contains a coding region for a mitochondrial protein which does not appear to be translated. Four functional domains were found in the PHO4 protein, which consists of 312 amino acid (aa) residues as deduced from the open reading frame of PHO4. A gel retardation assay with beta-galactosidase::PHO4 fused protein revealed that the 85-aa C terminus is the domain responsible for binding to the promoter DNA of PHO5, a gene under the control of PHO4. This region has similarities with the amphipathic helix-loop-helix motif of c-myc protein. Determination of the nucleotide sequences of four PHO4c mutant alleles and insertion and deletion analyses of PHO4 DNA indicated that a region from aa 163 to 202 is involved in interaction with a negative regulatory factor PHO80. Complementation of a pho4 null allele with the modified PHO4 DNAs suggested that the N-terminal region (1 to 109 aa), which is rich in acidic aa, is the transcriptional activation domain. The deleterious effects of various PHO4 mutations on the constitutive transcription of PHO5 in PHO4c mutant cells suggested that the region from aa 203 to 227 is involved in oligomerization of the PHO4 protein. (3)
Binding of transcription factors to DNA is a key regulatory step in the control of gene expression. DNA sequences with high affinity for transcription factors occur more frequently in the genome than instances of genes bound or regulated by these factors. Although several mechanisms have been identified that influence the specificity of transcriptional regulation, it is not known if these can explain the observed genome-wide pattern of binding or regulation for a given transcription factor.(4)
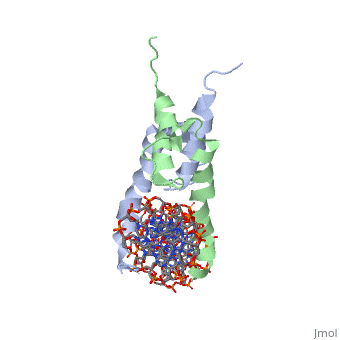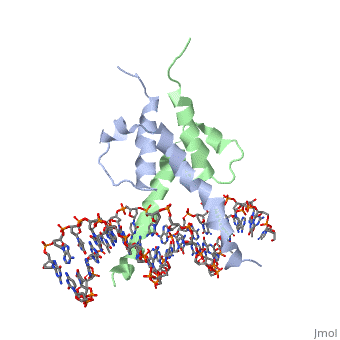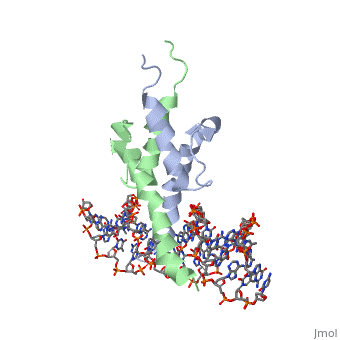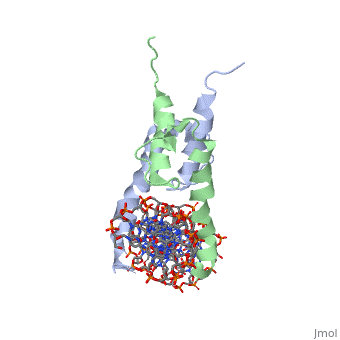
The crystal structure of a DNA-binding domain of PHO4 complexed with DNA at 2.8 A resolution revealed that the domain folds into a basic-helix-loop-helix (bHLH) motif with a long but compact loop that contains a short alpha-helical segment. This helical structure positions a tryptophan residue into an aromatic cluster so as to make the loop compact. PHO4 binds to DNA as a homodimer with direct reading of both the core E-box sequence CACGTG and its 3'-flanking bases. The 3'-flanking bases GG are recognized by Arg2 and His5. The residues involved in the E-box recognition are His5, Glu9 and Arg13, as already reported for bHLH/Zip proteins MAX and USF, and are different from those recognized by bHLH proteins MyoD and E47, although PHO4 is a bHLH protein.
Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition., Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T, EMBO J. 1997 Aug 1;16(15):4689-97. PMID:9303313
From MEDLINE®/PubMed®, a database of the U.S. National Library of Medicine.
Primary Sequence of PolypeptidePrimary Sequence of Polypeptide
Pho4 contains DNA binding site is Chain C with Primary Sequence of DNA (5’-CTCACACGTGGGACTAG-3’)
Primary Sequence of Polypeptide
MKRESHKHAEQARRNRLAVALHELASLIPAEWKQQNVSAAPSKATTVEAACRYIRHLQQNGST
1-MET-LYS-ARG-GLU-SER-HIS-LYS-HIS-ALA-GLU-GLN-ALA-ARG-ARG-ASN-ARG-LEU-ALA-VAL-ALA-LEU-HIS-GLU-LEU-ALA-SER-LEU-ILE-PRO-ALA-GLU-TRP-LYS-GLN-GLN-ASN-VAL-SER-ALA-ALA-PRO-SER-LYS-ALA-THR-THR-VAL-GLU-ALA-ALA-CYS-ARG-TYR-ILE-ARG-HIS-LEU-GLN-GLN-ASN-GLY-end File:Https://lh3.ggpht.com/ c3v6Rba79A693iTJrAGw59OQHYy0qDZ3hFGG73nzdIlKlGiYGFz M 6mprKgeR8wj6of84=s139
About this StructureAbout this Structure
The crystal structure of a DNA-binding domain of PHO4 complexed has DNA resolution of 2.8 A. This domain folds into a basic-helix-loop-helix (bHLH) motif with a long loop. The compact loop contains a short alpha-helical segment. This helical segment places a tryptophan residue into an aromatic cluster forming the compact loop.A homodimer is formed when PHO4 binds to DNA this complex has the ability to direct reading of both the core E-box sequence CACGTG and its 3'-flanking bases. The Arg2 and His5 are both able to recognize the 3'-flanking bases GG. The residues involved in the E-box recognition are His5, Glu9 and Arg13, as already reported for bHLH/Zip proteins MAX and USF, and are different from those recognized by bHLH proteins MyoD and E47, although PHO4 is a bHLH protein.
1a0a is a 4 chain structure with sequence from Saccharomyces cerevisiae. Full crystallographic information is available from OCA.
Predicted functional partners of Pho4
•PH0 80 •PHO85 •PHO2 •PHO81 •MSN5 •PSE1 •SUA7 •SPT 15 •PHO4 •IN04
Sequence comparison to large number of mammalian and Drophilia BHLH proteins reveales similarity in the bHLH region.
ReferenceReference
- ↑ Shimizu T, Toumoto A, Ihara K, Shimizu M, Kyogoku Y, Ogawa N, Oshima Y, Hakoshima T. Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition. EMBO J. 1997 Aug 1;16(15):4689-97. PMID:9303313 doi:10.1093/emboj/16.15.4689