G18secL03Tpc4
|
Outer surface protein C (OspC) of Borrelia burgdorferi is one of the major antigens on the surface of the Lyme disease spirochete, Borrelia burgdorferi, along with other outer surface proteins A and B (OspA and OspB, respectively). It greatly differs from OspA and OspB in both structure and function. The uniqueness of OspC is that it comes into play when the pathogen is being transmitted to humans or other mammals.OspC is critical for survival in or transmission to the tick or mammalian host.[1] OspC is being produced by Borrelia burgdorferi during a very short time interval when infected ticks start feeding, but its synthesis is known to slow down greatly after transmission to a mammalian host. It was demonstrated that those spirochetes that lack OspC are capable to replicate inside and migrate to the salivary glands of the tick vector but do not infect mammals. [2] Without OspC the spirochetes are believed to be unable to adapt to the environment inside the host. Therefore, OspC is believed to determine virulence of the spirochete to mammals, including humans.
Basic Structure DescriptionBasic Structure Description
- Primary Structure
The ospC gene is located on a 27 kb circular plasmid and encodes a lipoprotein of 22–23 kDa.[3] The protein is initially synthesized with an 18-amino-acid-long signal sequence which is removed during processing and lipidation at the amino proximal Cys residue. OspC proteins are highly polymorphic and this variability extends even to strains collected from a single geographical area.[4]
- Secondary Structure
OspC is predominantly α-helical in its secondary structure. β-sheets are also present, but they are rather short and not promininent.
- Tertiary Structure
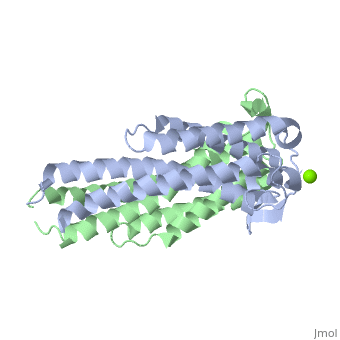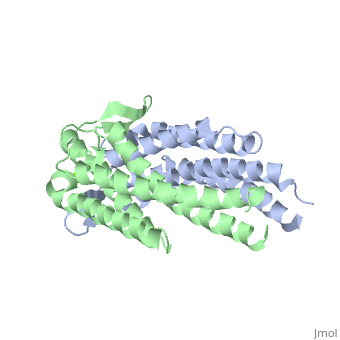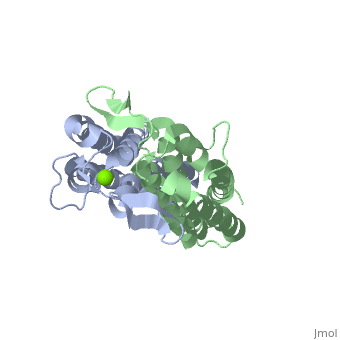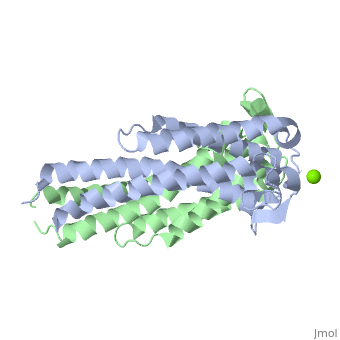
A single OspC monomer subunit is composed of 4 long and 1 short α-helices. Also, 2 short segments of β-sheets are observed near the binding site of the molecule.
- Quaternary Structure
OspC is a dimerized molecule, with 2 identical monomeric subunits comprising a dimer. However, for a binding event to occur, 2 dimers are necessary.
Major Hypothesized FunctionsMajor Hypothesized Functions
- Adaptation and survival in different host environments
Borrelia burgdorferi expresses OspA but not OspC when residing in the midgut of unfed ticks. However, when the tick starts feeding on mammals, OspC synthesis is induced and OspA is repressed.The switch is in part due to the change in temperature; OspC is induced at 32–37°C, but not at 24°C, and this upregulation is at the transcriptional and translational levels. Evidence suggests co-regulation of these two genes at the mRNA level. Clearly, to survive in both hosts, spirochetes have evolved mechanisms for sensing the different host environments and responding accordingly.[5]
- Binding and attachment to host's tissues
OspC may possibly be a binding protein contributing to a fundamental biological process and determining virulence of the bacteria. Several studies have shown that B.burgdorferi has a predilection for collagenous tissue and can interact with fibronectin and cellular collagens. The spirochetes can bind to a number of different cell types, including fibroblasts. Borrelia burgdorferi can bind to a novel circulating fibroblast-like cell called the peripheral blood fibrocyte, which expresses collagen types I and III as well as fibronectin, in a process that does not require OspA or OspB.[6]
Putative Binding SitePutative Binding Site
The binding site of OspC is believed to be located on the surface that projects away from the membrane and has a region with strong negative electrostatic potential. Cavities are formed at the top of the molecule away from the membrane surface. Each cavity has a volume of 50 Å3 and is formed by residues Ala75, Ile76, Gly77, Lys78, Lys79, Glu89, Ala90, Asp91, His92 and Asn93 of one monomer, and Gly94, Ser95, Ser98, Gly146, Lys147 and Glu148 of the other monomer.[7] Positively charged Magnesium ion Mg2+ between the two dimers demonstrates the location of hypothesized binding site.
Role of OspC in Lyme DiseaseRole of OspC in Lyme Disease
OspC-based Vaccine Against Lyme DiseaseOspC-based Vaccine Against Lyme Disease
Researchers believe that an OspC based vaccine would be more effective against Lyme disease than the current OspA based vaccine once further research is conducted regarding the three dimensional structure of OspC's, especially the structure of invasive strains [8]. An OspC vaccine would have significant advantages over the current OspA vaccine, but there are several problems that require further research before the vaccine can be made.
Main Advantages of Developing OspC-based VaccineMain Advantages of Developing OspC-based Vaccine
Unlike OspC, the OspA protein is only present in the Borrelia burgdorferi while they are in the midgut of the cold blooded tick, and not in the host. Once the tick begins to feed on its warm blooded mammalian host, the Borrelia burgdorferi migrate from the midgut of the tick to the salivary glands and OspC is produced in the host's bloodstream. [9] Because of this, when a host is vaccinated with the OspA vaccine, antibodies to the OspA protein can only kill the bacteria inside of the tick if it ingests the antibodies during feeding. [10] If the bacteria enter the host, it can differentiate into several forms for which the vaccine cannot protect against. In contrast, an OspC based vaccine would allow the host to make antibodies to kill the Borrelia burgdorferi after they enter the host's body. [11]
Main Problems with Application of OspC-based VaccineMain Problems with Application of OspC-based Vaccine
Notes and References
- ↑ Stewart PE, Thalken R, Bono JL, Rosa P. Isolation of a circular plasmid region sufficient for autonomous replication and transformation of infectious Borrelia burgdorferi. Mol Microbiol. 2001 Feb;39(3):714-21. PMID:11169111
- ↑ D. Kumaran1, S. Eswaramoorthy1, B.J. Luft2, S. Koide3, J.J. Dunn1, C.L. Lawson1,4 and S. Swaminathan1. Crystal structure of outer surface protein C (OspC) from the Lyme disease spirochete, Borrelia burgdorferi.The EMBO Journal (2001) 20, 971 - 978 [1]
- ↑ Marconi RT, Samuels DS, Garon CF. Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J Bacteriol. 1993 Feb;175(4):926-32. PMID:7679385
- ↑ D. Kumaran1, S. Eswaramoorthy1, B.J. Luft2, S. Koide3, J.J. Dunn1, C.L. Lawson1,4 and S. Swaminathan1. Crystal structure of outer surface protein C (OspC) from the Lyme disease spirochete, Borrelia burgdorferi.The EMBO Journal (2001) 20, 971 - 978 [2]
- ↑ D. Kumaran1, S. Eswaramoorthy1, B.J. Luft2, S. Koide3, J.J. Dunn1, C.L. Lawson1,4 and S. Swaminathan1. Crystal structure of outer surface protein C (OspC) from the Lyme disease spirochete, Borrelia burgdorferi.The EMBO Journal (2001) 20, 971 - 978 [3]
- ↑ Grab DJ, Lanners H, Martin LN, Chesney J, Cai C, Adkisson HD, Bucala R. Interaction of Borrelia burgdorferi with peripheral blood fibrocytes, antigen-presenting cells with the potential for connective tissue targeting. Mol Med. 1999 Jan;5(1):46-54. PMID:10072447
- ↑ D. Kumaran1, S. Eswaramoorthy1, B.J. Luft2, S. Koide3, J.J. Dunn1, C.L. Lawson1,4 and S. Swaminathan1. Crystal structure of outer surface protein C (OspC) from the Lyme disease spirochete, Borrelia burgdorferi.The EMBO Journal (2001) 20, 971 - 978 [4]
- ↑ D. Kumaran1, S. Eswaramoorthy1, B.J. Luft2, S. Koide3, J.J. Dunn1, C.L. Lawson1,4 and S. Swaminathan1. Crystal structure of outer surface protein C (OspC) from the Lyme disease spirochete, Borrelia burgdorferi.The EMBO Journal (2001) 20, 971 - 978 [5]
- ↑ D. Greenburg, M.S. Rowe. Brookhaven Scientists Determine Key Lime Disease Protein Structure.Brookhaven National Laboratory (2001)[6]
- ↑ D. Greenburg, M.S. Rowe. Brookhaven Scientists Determine Key Lime Disease Protein Structure.Brookhaven National Laboratory (2001)[7]
- ↑ D. Greenburg, M.S. Rowe. Brookhaven Scientists Determine Key Lime Disease Protein Structure.Brookhaven National Laboratory (2001)[8]